Human-specific transcriptional networks in the brain
- PMID: 22920253
- PMCID: PMC3645834
- DOI: 10.1016/j.neuron.2012.05.034
Human-specific transcriptional networks in the brain
Abstract
Understanding human-specific patterns of brain gene expression and regulation can provide key insights into human brain evolution and speciation. Here, we use next-generation sequencing, and Illumina and Affymetrix microarray platforms, to compare the transcriptome of human, chimpanzee, and macaque telencephalon. Our analysis reveals a predominance of genes differentially expressed within human frontal lobe and a striking increase in transcriptional complexity specific to the human lineage in the frontal lobe. In contrast, caudate nucleus gene expression is highly conserved. We also identify gene coexpression signatures related to either neuronal processes or neuropsychiatric diseases, including a human-specific module with CLOCK as its hub gene and another module enriched for neuronal morphological processes and genes coexpressed with FOXP2, a gene important for language evolution. These data demonstrate that transcriptional networks have undergone evolutionary remodeling even within a given brain region, providing a window through which to view the foundation of uniquely human cognitive capacities.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

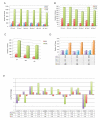

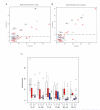
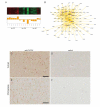


Comment in
-
Brain, know thy transcriptome, know thyself.Neuron. 2012 Aug 23;75(4):543-5. doi: 10.1016/j.neuron.2012.08.005. Neuron. 2012. PMID: 22920245
-
Neurogenetics: What makes a human brain?Nat Rev Neurosci. 2012 Oct;13(10):655. doi: 10.1038/nrn3355. Nat Rev Neurosci. 2012. PMID: 22992645 No abstract available.
References
-
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci U S A. 1999;96:9885–9890. - PMC - PubMed
-
- Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K., Erwin, J., Park S, Goubert V, Hof PR. The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct Funct. 2010;214:495–517. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

