Comparative metagenomics of two microbial mats at Cuatro Ciénegas Basin II: community structure and composition in oligotrophic environments
- PMID: 22920516
- PMCID: PMC3426889
- DOI: 10.1089/ast.2011.0724
Comparative metagenomics of two microbial mats at Cuatro Ciénegas Basin II: community structure and composition in oligotrophic environments
Abstract
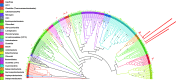
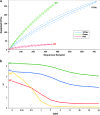
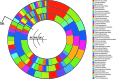
Microbial mats are self-sustained, functionally complex ecosystems that make good models for the understanding of past and present microbial ecosystems as well as putative extraterrestrial ecosystems. Ecological theory suggests that the composition of these communities might be affected by nutrient availability and disturbance frequency. We characterized two microbial mats from two contrasting environments in the oligotrophic Cuatro Ciénegas Basin: a permanent green pool and a red desiccation pond. We analyzed their taxonomic structure and composition by means of 16S rRNA clone libraries and metagenomics and inferred their metabolic role by the analysis of functional traits in the most abundant organisms. Both mats showed a high diversity with metabolically diverse members and strongly differed in structure and composition. The green mat had a higher species richness and evenness than the red mat, which was dominated by a lineage of Pseudomonas. Autotrophs were abundant in the green mat, and heterotrophs were abundant in the red mat. When comparing with other mats and stromatolites, we found that taxonomic composition was not shared at species level but at order level, which suggests environmental filtering for phylogenetically conserved functional traits with random selection of particular organisms. The highest diversity and composition similarity was observed among systems from stable environments, which suggests that disturbance regimes might affect diversity more strongly than nutrient availability, since oligotrophy does not appear to prevent the establishment of complex and diverse microbial mat communities. These results are discussed in light of the search for extraterrestrial life.
Figures


References
-
- Abed R.M. Kohls K. de Beer D. Effect of salinity changes on the bacterial diversity, photosynthesis and oxygen consumption of cyanobacterial mats from an intertidal flat of the Arabian Gulf. Environ Microbiol. 2007;9:1384–1392. - PubMed
-
- Allen M.A. Goh F. Burns B.P. Neilan B.A. Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology. 2009;7:82–96. - PubMed
-
- Allwood A.C. Walter M.R. Kamber B.S. Marshall C.P. Burch I.W. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

