Dissecting the role of low-complexity regions in the evolution of vertebrate proteins
- PMID: 22920595
- PMCID: PMC3523016
- DOI: 10.1186/1471-2148-12-155
Dissecting the role of low-complexity regions in the evolution of vertebrate proteins
Abstract
Background: Low-complexity regions (LCRs) in proteins are tracts that are highly enriched in one or a few amino acids. Given their high abundance, and their capacity to expand in relatively short periods of time through replication slippage, they can greatly contribute to increase protein sequence space and generate novel protein functions. However, little is known about the global impact of LCRs on protein evolution.
Results: We have traced back the evolutionary history of 2,802 LCRs from a large set of homologous protein families from H.sapiens, M.musculus, G.gallus, D.rerio and C.intestinalis. Transcriptional factors and other regulatory functions are overrepresented in proteins containing LCRs. We have found that the gain of novel LCRs is frequently associated with repeat expansion whereas the loss of LCRs is more often due to accumulation of amino acid substitutions as opposed to deletions. This dichotomy results in net protein sequence gain over time. We have detected a significant increase in the rate of accumulation of novel LCRs in the ancestral Amniota and mammalian branches, and a reduction in the chicken branch. Alanine and/or glycine-rich LCRs are overrepresented in recently emerged LCR sets from all branches, suggesting that their expansion is better tolerated than for other LCR types. LCRs enriched in positively charged amino acids show the contrary pattern, indicating an important effect of purifying selection in their maintenance.
Conclusion: We have performed the first large-scale study on the evolutionary dynamics of LCRs in protein families. The study has shown that the composition of an LCR is an important determinant of its evolutionary pattern.
Figures

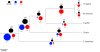
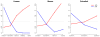
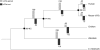
References
-
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

