H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells
- PMID: 22920947
- PMCID: PMC3473242
- DOI: 10.1186/1471-2164-13-424
H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells
Abstract
Background: Transcription regulation in pluripotent embryonic stem (ES) cells is a complex process that involves multitude of regulatory layers, one of which is post-translational modification of histones. Acetylation of specific lysine residues of histones plays a key role in regulating gene expression.
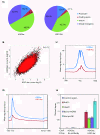
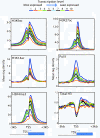
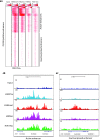
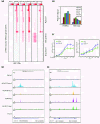
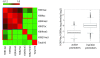
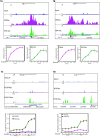
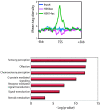
Results: Here we have investigated the genome-wide occurrence of two histone marks, acetylation of histone H3K9 and K14 (H3K9ac and H3K14ac), in mouse embryonic stem (mES) cells. Genome-wide H3K9ac and H3K14ac show very high correlation between each other as well as with other histone marks (such as H3K4me3) suggesting a coordinated regulation of active histone marks. Moreover, the levels of H3K9ac and H3K14ac directly correlate with the CpG content of the promoters attesting the importance of sequences underlying the specifically modified nucleosomes. Our data provide evidence that H3K9ac and H3K14ac are also present over the previously described bivalent promoters, along with H3K4me3 and H3K27me3. Furthermore, like H3K27ac, H3K9ac and H3K14ac can also differentiate active enhancers from inactive ones. Although, H3K9ac and H3K14ac, a hallmark of gene activation exhibit remarkable correlation over active and bivalent promoters as well as distal regulatory elements, a subset of inactive promoters is selectively enriched for H3K14ac.
Conclusions: Our study suggests that chromatin modifications, such as H3K9ac and H3K14ac, are part of the active promoter state, are present over bivalent promoters and active enhancers and that the extent of H3K9 and H3K14 acetylation could be driven by cis regulatory elements such as CpG content at promoters. Our study also suggests that a subset of inactive promoters is selectively and specifically enriched for H3K14ac. This observation suggests that histone acetyl transferases (HATs) prime inactive genes by H3K14ac for stimuli dependent activation. In conclusion our study demonstrates a wider role for H3K9ac and H3K14ac in gene regulation than originally thought.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

