Functional analysis of family GH36 α-galactosidases from Ruminococcus gnavus E1: insights into the metabolism of a plant oligosaccharide by a human gut symbiont
- PMID: 22923411
- PMCID: PMC3485720
- DOI: 10.1128/AEM.01350-12
Functional analysis of family GH36 α-galactosidases from Ruminococcus gnavus E1: insights into the metabolism of a plant oligosaccharide by a human gut symbiont
Abstract
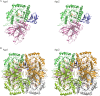
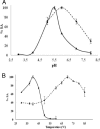
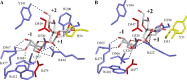
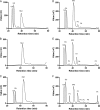
Ruminococcus gnavus belongs to the 57 most common species present in 90% of individuals. Previously, we identified an α-galactosidase (Aga1) belonging to glycoside hydrolase (GH) family 36 from R. gnavus E1 (M. Aguilera, H. Rakotoarivonina, A. Brutus, T. Giardina, G. Simon, and M. Fons, Res. Microbiol. 163:14-21, 2012). Here, we identified a novel GH36-encoding gene from the same strain and termed it aga2. Although aga1 showed a very simple genetic organization, aga2 is part of an operon of unique structure, including genes putatively encoding a regulator, a GH13, two phosphotransferase system (PTS) sequences, and a GH32, probably involved in extracellular and intracellular sucrose assimilation. The 727-amino-acid (aa) deduced Aga2 protein shares approximately 45% identity with Aga1. Both Aga1 and Aga2 expressed in Escherichia coli showed strict specificity for α-linked galactose. Both enzymes were active on natural substrates such as melibiose, raffinose, and stachyose. Aga1 and Aga2 occurred as homotetramers in solution, as shown by analytical ultracentrifugation. Modeling of Aga1 and Aga2 identified key amino acids which may be involved in substrate specificity and stabilization of the α-linked galactoside substrates within the active site. Furthermore, Aga1 and Aga2 were both able to perform transglycosylation reactions with α-(1,6) regioselectivity, leading to the formation of product structures up to [Hex](12) and [Hex](8), respectively. We suggest that Aga1 and Aga2 play essential roles in the metabolism of dietary oligosaccharides and could be used for the design of galacto-oligosaccharide (GOS) prebiotics, known to selectively modulate the beneficial gut microbiota.
Figures



References
-
- Aguilera M, et al. 2012. Aga1, the first alpha-galactosidase from the human bacteria Ruminococcus gnavus E1, efficiently transcribed in gut conditions. Res. Microbiol. 163:14–21 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

