Identification of molecular pathway aberrations in uterine serous carcinoma by genome-wide analyses
- PMID: 22923510
- PMCID: PMC3692380
- DOI: 10.1093/jnci/djs345
Identification of molecular pathway aberrations in uterine serous carcinoma by genome-wide analyses
Abstract
Background: Uterine cancer is the fourth most common malignancy in women, and uterine serous carcinoma is the most aggressive subtype. However, the molecular pathogenesis of uterine serous carcinoma is largely unknown. We analyzed the genomes of uterine serous carcinoma samples to better understand the molecular genetic characteristics of this cancer.
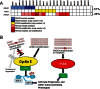
Methods: Whole-exome sequencing was performed on 10 uterine serous carcinomas and the matched normal blood or tissue samples. Somatically acquired sequence mutations were further verified by Sanger sequencing. The most frequent molecular genetic changes were further validated by Sanger sequencing in 66 additional uterine serous carcinomas and in nine serous endometrial intraepithelial carcinomas (the preinvasive precursor of uterine serous carcinoma) that were isolated by laser capture microdissection. In addition, gene copy number was characterized by single-nucleotide polymorphism (SNP) arrays in 23 uterine serous carcinomas, including 10 that were subjected to whole-exome sequencing.
Results: We found frequent somatic mutations in TP53 (81.6%), PIK3CA (23.7%), FBXW7 (19.7%), and PPP2R1A (18.4%) among the 76 uterine serous carcinomas examined. All nine serous carcinomas that had an associated serous endometrial intraepithelial carcinoma had concordant PIK3CA, PPP2R1A, and TP53 mutation status between uterine serous carcinoma and the concurrent serous endometrial intraepithelial carcinoma component. DNA copy number analysis revealed frequent genomic amplification of the CCNE1 locus (which encodes cyclin E, a known substrate of FBXW7) and deletion of the FBXW7 locus. Among 23 uterine serous carcinomas that were subjected to SNP array analysis, seven tumors with FBXW7 mutations (four tumors with point mutations, three tumors with hemizygous deletions) did not have CCNE1 amplification, and 13 (57%) tumors had either a molecular genetic alteration in FBXW7 or CCNE1 amplification. Nearly half of these uterine serous carcinomas (48%) harbored PIK3CA mutation and/or PIK3CA amplification.
Conclusion: Molecular genetic aberrations involving the p53, cyclin E-FBXW7, and PI3K pathways represent major mechanisms in the development of uterine serous carcinoma.
Figures


Similar articles
-
Frequent CCNE1 amplification in endometrial intraepithelial carcinoma and uterine serous carcinoma.Mod Pathol. 2014 Jul;27(7):1014-9. doi: 10.1038/modpathol.2013.209. Epub 2013 Dec 6. Mod Pathol. 2014. PMID: 24309323
-
Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations.Mod Pathol. 2015 Apr;28(4):505-14. doi: 10.1038/modpathol.2014.143. Epub 2014 Nov 14. Mod Pathol. 2015. PMID: 25394778
-
The FOXA2 transcription factor is frequently somatically mutated in uterine carcinosarcomas and carcinomas.Cancer. 2018 Jan 1;124(1):65-73. doi: 10.1002/cncr.30971. Epub 2017 Sep 21. Cancer. 2018. PMID: 28940304 Free PMC article.
-
Novel targeted therapies in uterine serous carcinoma, an aggressive variant of endometrial cancer.Discov Med. 2016 Apr;21(116):293-303. Discov Med. 2016. PMID: 27232515 Review.
-
Targeted therapy in uterine serous carcinoma: an aggressive variant of endometrial cancer.Womens Health (Lond). 2014 Jan;10(1):45-57. doi: 10.2217/whe.13.72. Womens Health (Lond). 2014. PMID: 24328598 Free PMC article. Review.
Cited by
-
Ligand-directed targeting of lymphatic vessels uncovers mechanistic insights in melanoma metastasis.Proc Natl Acad Sci U S A. 2015 Feb 24;112(8):2521-6. doi: 10.1073/pnas.1424994112. Epub 2015 Feb 6. Proc Natl Acad Sci U S A. 2015. PMID: 25659743 Free PMC article.
-
DNA Methylation Module Network-Based Prognosis and Molecular Typing of Cancer.Genes (Basel). 2019 Jul 28;10(8):571. doi: 10.3390/genes10080571. Genes (Basel). 2019. PMID: 31357729 Free PMC article.
-
Molecular Evaluation of Endometrial Dedifferentiated Carcinoma, Endometrioid Carcinoma, Carcinosarcoma, and Serous Carcinoma Using a Custom-Made Small Cancer Panel.Pathol Oncol Res. 2021 Dec 23;27:1610013. doi: 10.3389/pore.2021.1610013. eCollection 2021. Pathol Oncol Res. 2021. PMID: 35002543 Free PMC article.
-
miR-137 is a tumor suppressor in endometrial cancer and is repressed by DNA hypermethylation.Lab Invest. 2018 Nov;98(11):1397-1407. doi: 10.1038/s41374-018-0092-x. Epub 2018 Jun 28. Lab Invest. 2018. PMID: 29955087 Free PMC article.
-
The impact of microRNA-mediated PI3K/AKT signaling on epithelial-mesenchymal transition and cancer stemness in endometrial cancer.J Transl Med. 2014 Aug 21;12:231. doi: 10.1186/s12967-014-0231-0. J Transl Med. 2014. PMID: 25141911 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

