Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing
- PMID: 22923523
- PMCID: PMC3488221
- DOI: 10.1093/nar/gks753
Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing
Abstract
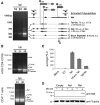
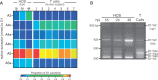
Alternative RNA splicing greatly expands the repertoire of proteins encoded by genomes. Next-generation sequencing (NGS) is attractive for studying alternative splicing because of the efficiency and low cost per base, but short reads typical of NGS only report mRNA fragments containing one or few splice junctions. Here, we used single-molecule amplification and long-read sequencing to study the HIV-1 provirus, which is only 9700 bp in length, but encodes nine major proteins via alternative splicing. Our data showed that the clinical isolate HIV-1(89.6) produces at least 109 different spliced RNAs, including a previously unappreciated ∼1 kb class of messages, two of which encode new proteins. HIV-1 message populations differed between cell types, longitudinally during infection, and among T cells from different human donors. These findings open a new window on a little studied aspect of HIV-1 replication, suggest therapeutic opportunities and provide advanced tools for the study of alternative splicing.
Figures


References
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. - PubMed
-
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. - PubMed

