Detection of alternative lengthening of telomeres by telomere quantitative PCR
- PMID: 22923525
- PMCID: PMC3553966
- DOI: 10.1093/nar/gks781
Detection of alternative lengthening of telomeres by telomere quantitative PCR
Abstract
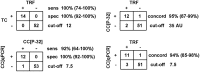
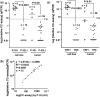
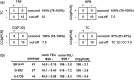
Alternative lengthening of telomeres (ALT) is one of the two known telomere length maintenance mechanisms that are essential for the unlimited proliferation potential of cancer cells. Existing methods for detecting ALT in tumors require substantial amounts of tumor material and are labor intensive, making it difficult to study prevalence and prognostic significance of ALT in large tumor cohorts. Here, we present a novel strategy utilizing telomere quantitative PCR to diagnose ALT. The protocol is more rapid than conventional methods and scrutinizes two distinct characteristics of ALT cells concurrently: long telomeres and the presence of C-circles (partially double-stranded circles of telomeric C-strand DNA). Requiring only 30 ng of genomic DNA, this protocol will facilitate large-scale studies of ALT in tumors and can be readily adopted by clinical laboratories.
Figures



References
-
- Reddel RR. The role of senescence and immortalization in carcinogenesis. Carcinogenesis. 2000;21:477–484. - PubMed
-
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. - PubMed
-
- Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. - PubMed
-
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. - PubMed
-
- Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 2005;11:217–225. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

