Engineered synthetic polymer nanoparticles as IgG affinity ligands
- PMID: 22924890
- PMCID: PMC3482410
- DOI: 10.1021/ja303612d
Engineered synthetic polymer nanoparticles as IgG affinity ligands
Abstract
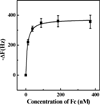
A process for the preparation of an abiotic protein affinity ligand is described. The affinity ligand, a synthetic polymer hydrogel nanoparticle (NP), is formulated with functional groups complementary to the surface presentation of the target protein. An iterative process is used to improve affinity by optimizing the composition and proportion of functional monomers. Since the polymer NPs are formed by a kinetically driven process, the sequence of functional monomers in the polymer chain is not controlled; only the average composition can be adjusted by the stoichiometry of the monomers in the feed. To compensate for this the hydrogel NP is lightly cross-linked resulting in chain flexibility that takes place on a submillisecond time scale allowing the polymer to "map" onto a protein surface with complementary functionality. In this study, we report a lightly cross-linked (2%) N-isopropyl acrylamide (NIPAm) synthetic polymer NP (50-65 nm) incorporating hydrophobic and carboxylate groups that binds with high affinity to the Fc fragment of IgG. The affinity and amount of NP bound to IgG is pH dependent. The hydrogel NP inhibits protein A binding to the Fc domain at pH 5.5, but not at pH 7.3. A computational analysis was used to identify potential NP-protein interaction sites. Candidates include a NP binding domain that overlaps with the protein A-Fc binding domain at pH 5.5. The computational analysis supports the inhibition experimental results and is attributed to the difference in the charged state of histidine residues. Affinity of the NP (3.5-8.5 nM) to the Fc domain at pH 5.5 is comparable to protein A at pH 7. These results establish that engineered synthetic polymer NPs can be formulated with an intrinsic affinity to a specific domain of a large biomacromolecule.
Figures
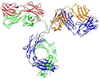








References
-
- Meenach SA, Hilt JZ, Anderson KW. Acta Biomater. 2010;6:1039–1046. - PubMed
-
- Tong R, Christian DA, Tang L, Cabral H, Baker JR, Kataoka K, Discher DE, Cheng JJ. Mrs Bull. 2009;34:422–431.
-
- Messerschmidt SKE, Musyanovych A, Altvater M, Scheurich P, Pfizenmaier K, Landfester K, Kontermann RE. J. Control. Release. 2009;137:69–77. - PubMed
-
- Davis ME, Chen Z, Shin DM. Nat. Rev. Drug Discov. 2008;7:771–782. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

