Characterization of CDKN2A(p16) methylation and impact in colorectal cancer: systematic analysis using pyrosequencing
- PMID: 22925370
- PMCID: PMC3479016
- DOI: 10.1186/1479-5876-10-173
Characterization of CDKN2A(p16) methylation and impact in colorectal cancer: systematic analysis using pyrosequencing
Abstract
Background: The aim of this study is to analyse CDKN2A methylation using pyrosequencing on a large cohort of colorectal cancers and corresponding non-neoplastic tissues. In a second step, the effect of methylation on clinical outcome is addressed.
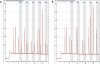
Methods: Primary colorectal cancers and matched non-neoplastic tissues from 432 patients underwent CDKN2A methylation analysis by pyrosequencing (PyroMarkQ96). Methylation was then related to clinical outcome, microsatellite instability (MSI), and BRAF and KRAS mutation. Different amplification conditions (35 to 50 PCR cycles) using a range of 0-100% methylated DNA were tested.
Results: Background methylation was at most 10% with ≥35 PCR cycles. Correlation of observed and expected values was high, even at low methylation levels (0.02%, 0.6%, 2%). Accuracy of detection was optimal with 45 PCR cycles. Methylation in normal mucosa ranged from 0 to >90% in some cases. Based on the maximum value of 10% background, positivity was defined as a ≥20% difference in methylation between tumor and normal tissue, which occurred in 87 cases. CDKN2A methylation positivity was associated with MSI (p = 0.025), BRAF mutation (p < 0.0001), higher tumor grade (p < 0.0001), mucinous histology (p = 0.0209) but not with KRAS mutation. CDKN2A methylation had an independent adverse effect (p = 0.0058) on prognosis.
Conclusion: The non-negligible CDKN2A methylation of normal colorectal mucosa may confound the assessment of tumor-specific hypermethylation, suggesting that corresponding non-neoplastic tissue should be used as a control. CDKN2A methylation is robustly detected by pyrosequencing, even at low levels, suggesting that this unfavorable prognostic biomarker warrants investigation in prospective studies.
Figures


References
-
- Carragher LA, Snell KR, Giblett SM, Aldridge VS, Patel B, Cook SJ, Winton DJ, Marais R, Pritchard CA. V600EBraf induces gastrointestinal crypt senescence and promotes tumour progression through enhanced CpG methylation of p16INK4a. EMBO Mol Med. 2010;2:458–471. doi: 10.1002/emmm.201000099. - DOI - PMC - PubMed
-
- Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer. 2011;128:1080–1094. doi: 10.1002/ijc.25432. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

