Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei
- PMID: 22925482
- PMCID: PMC3447711
- DOI: 10.1186/1471-2164-13-427
Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei
Abstract
Background: At the core of the RNA interference (RNAi) pathway in Trypanosoma brucei is a single Argonaute protein, TbAGO1, with an established role in controlling retroposon and repeat transcripts. Recent evidence from higher eukaryotes suggests that a variety of genomic sequences with the potential to produce double-stranded RNA are sources for small interfering RNAs (siRNAs).
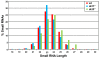
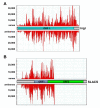
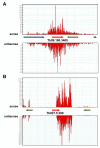
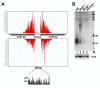
Results: To test whether such endogenous siRNAs are present in T. brucei and to probe the individual role of the two Dicer-like enzymes, we affinity purified TbAGO1 from wild-type procyclic trypanosomes, as well as from cells deficient in the cytoplasmic (TbDCL1) or nuclear (TbDCL2) Dicer, and subjected the bound RNAs to Illumina high-throughput sequencing. In wild-type cells the majority of reads originated from two classes of retroposons. We also considerably expanded the repertoire of trypanosome siRNAs to encompass a family of 147-bp satellite-like repeats, many of the regions where RNA polymerase II transcription converges, large inverted repeats and two pseudogenes. Production of these newly described siRNAs is strictly dependent on the nuclear DCL2. Notably, our data indicate that putative centromeric regions, excluding the CIR147 repeats, are not a significant source for endogenous siRNAs.
Conclusions: Our data suggest that endogenous RNAi targets may be as evolutionarily old as the mechanism itself.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

