Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage
- PMID: 22925495
- PMCID: PMC3491373
- DOI: 10.1186/gb-2012-13-8-r74
Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage
Abstract
Background: Bathycoccus prasinos is an extremely small cosmopolitan marine green alga whose cells are covered with intricate spider's web patterned scales that develop within the Golgi cisternae before their transport to the cell surface. The objective of this work is to sequence and analyze its genome, and to present a comparative analysis with other known genomes of the green lineage.
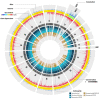
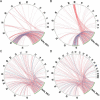
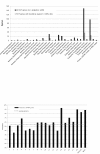
Research: Its small genome of 15 Mb consists of 19 chromosomes and lacks transposons. Although 70% of all B. prasinos genes share similarities with other Viridiplantae genes, up to 428 genes were probably acquired by horizontal gene transfer, mainly from other eukaryotes. Two chromosomes, one big and one small, are atypical, an unusual synapomorphic feature within the Mamiellales. Genes on these atypical outlier chromosomes show lower GC content and a significant fraction of putative horizontal gene transfer genes. Whereas the small outlier chromosome lacks colinearity with other Mamiellales and contains many unknown genes without homologs in other species, the big outlier shows a higher intron content, increased expression levels and a unique clustering pattern of housekeeping functionalities. Four gene families are highly expanded in B. prasinos, including sialyltransferases, sialidases, ankyrin repeats and zinc ion-binding genes, and we hypothesize that these genes are associated with the process of scale biogenesis.
Conclusion: The minimal genomes of the Mamiellophyceae provide a baseline for evolutionary and functional analyses of metabolic processes in green plants.
Figures


Similar articles
-
Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer.BMC Genomics. 2008 Jun 16;9:290. doi: 10.1186/1471-2164-9-290. BMC Genomics. 2008. PMID: 18558012 Free PMC article.
-
The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales.BMC Biol. 2005 Oct 20;3:22. doi: 10.1186/1741-7007-3-22. BMC Biol. 2005. PMID: 16236178 Free PMC article.
-
Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants.BMC Genomics. 2016 Mar 31;17:267. doi: 10.1186/s12864-016-2585-6. BMC Genomics. 2016. PMID: 27029936 Free PMC article.
-
Genome diversity in the smallest marine photosynthetic eukaryotes.Res Microbiol. 2011 Jul-Aug;162(6):570-7. doi: 10.1016/j.resmic.2011.04.005. Epub 2011 Apr 21. Res Microbiol. 2011. PMID: 21540104 Review.
-
Gene content, organization and molecular evolution of plant organellar genomes and sex chromosomes: insights from the case of the liverwort Marchantia polymorpha.Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(3):108-24. doi: 10.2183/pjab.85.108. Proc Jpn Acad Ser B Phys Biol Sci. 2009. PMID: 19282647 Free PMC article. Review.
Cited by
-
Alga-PrAS (Algal Protein Annotation Suite): A Database of Comprehensive Annotation in Algal Proteomes.Plant Cell Physiol. 2017 Jan 1;58(1):e6. doi: 10.1093/pcp/pcw212. Plant Cell Physiol. 2017. PMID: 28069893 Free PMC article.
-
Diversity and oceanic distribution of prasinophytes clade VII, the dominant group of green algae in oceanic waters.ISME J. 2017 Feb;11(2):512-528. doi: 10.1038/ismej.2016.120. Epub 2016 Oct 25. ISME J. 2017. PMID: 27779617 Free PMC article.
-
Metabolomic Insights into Marine Phytoplankton Diversity.Mar Drugs. 2020 Jan 25;18(2):78. doi: 10.3390/md18020078. Mar Drugs. 2020. PMID: 31991720 Free PMC article.
-
Genomic characterisation and ecological distribution of Mantoniella tinhauana: a novel Mamiellophycean green alga from the Western Pacific.Front Microbiol. 2024 May 7;15:1358574. doi: 10.3389/fmicb.2024.1358574. eCollection 2024. Front Microbiol. 2024. PMID: 38774501 Free PMC article.
-
Marine Prasinoviruses and Their Tiny Plankton Hosts: A Review.Viruses. 2017 Mar 15;9(3):43. doi: 10.3390/v9030043. Viruses. 2017. PMID: 28294997 Free PMC article. Review.
References
-
- Field BC, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. - PubMed
-
- Li WKW. Primary productivity of prochlorophytes cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. doi: 10.4319/lo.1994.39.1.0169. - DOI
-
- Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–179. doi: 10.4319/lo.2004.49.1.0168. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

