3Drefine: consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization
- PMID: 22927229
- PMCID: PMC3634918
- DOI: 10.1002/prot.24167
3Drefine: consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization
Abstract
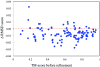
One of the major limitations of computational protein structure prediction is the deviation of predicted models from their experimentally derived true, native structures. The limitations often hinder the possibility of applying computational protein structure prediction methods in biochemical assignment and drug design that are very sensitive to structural details. Refinement of these low-resolution predicted models to high-resolution structures close to the native state, however, has proven to be extremely challenging. Thus, protein structure refinement remains a largely unsolved problem. Critical assessment of techniques for protein structure prediction (CASP) specifically indicated that most predictors participating in the refinement category still did not consistently improve model quality. Here, we propose a two-step refinement protocol, called 3Drefine, to consistently bring the initial model closer to the native structure. The first step is based on optimization of hydrogen bonding (HB) network and the second step applies atomic-level energy minimization on the optimized model using a composite physics and knowledge-based force fields. The approach has been evaluated on the CASP benchmark data and it exhibits consistent improvement over the initial structure in both global and local structural quality measures. 3Drefine method is also computationally inexpensive, consuming only few minutes of CPU time to refine a protein of typical length (300 residues). 3Drefine web server is freely available at http://sysbio.rnet.missouri.edu/3Drefine/.
Copyright © 2012 Wiley Periodicals, Inc.
Figures

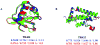


References
-
- Sali A, Blundell T. Comparative protein modelling by satisfaction of spatial restraints. Protein Structure by Distance Analysis. 1994;64:C86. - PubMed
-
- Fischer D. 3D-SHOTGUN: A novel, cooperative, fold-recognition meta-predictor. Proteins: Structure Function and Bioinformatics. 2003;51:434–441. - PubMed
-
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins: Structure Function and Bioinformatics. 2007;69:108–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

