Cytosolic sensing of extracellular self-DNA transported into monocytes by the antimicrobial peptide LL37
- PMID: 22927244
- PMCID: PMC3488884
- DOI: 10.1182/blood-2012-01-401364
Cytosolic sensing of extracellular self-DNA transported into monocytes by the antimicrobial peptide LL37
Abstract
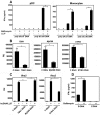
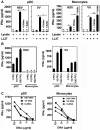
The intracellular location of nucleic acid sensors prevents recognition of extracellular self-DNA released by dying cells. However, on forming a complex with the endogenous antimicrobial peptide LL37, extracellular DNA is transported into endosomal compartments of plasmacytoid dendritic cells, leading to activation of Toll-like receptor-9 and induction of type I IFNs. Whether LL37 also transports self-DNA into nonplasmacytoid dendritic cells, leading to type I IFN production via other intracellular DNA receptors is unknown. Here we found that LL37 very efficiently transports self-DNA into monocytes, leading the production of type I IFNs in a Toll-like receptor-independent manner. This type I IFN induction was mediated by double-stranded B form DNA, regardless of its sequence, CpG content, or methylation status, and required signaling through the adaptor protein STING and TBK1 kinase, indicating the involvement of cytosolic DNA sensors. Thus, our study identifies a novel link between the antimicrobial peptides and type I IFN responses involving DNA-dependent activation of cytosolic sensors in monocytes.
Figures
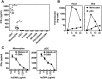
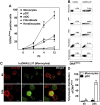
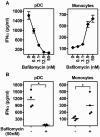
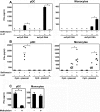
References
-
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autpimmunity. Annu Rev Immunol. 2005;23:307–336. - PubMed
-
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. - PubMed
-
- Luft T, Pang KC, Thomas E, et al. Type I IFNs enhance the terminal differentiation of dendritic cells. J Immunol. 1998;161(4):1947–1953. - PubMed
-
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10(2):123–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

