Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments
- PMID: 22927404
- PMCID: PMC3443165
- DOI: 10.1073/pnas.1207516109
Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments
Abstract
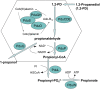
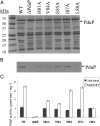
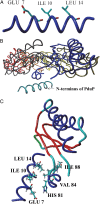
Bacterial microcompartments (MCPs) are a widespread family of proteinaceous organelles that consist of metabolic enzymes encapsulated within a protein shell. For MCPs to function specific enzymes must be encapsulated. We recently reported that a short N-terminal targeting sequence of propionaldehyde dehydrogenase (PduP) is necessary and sufficient for the packaging of enzymes into a MCP that functions in 1,2-propanediol (1,2-PD) utilization (Pdu) by Salmonella enterica. Here we show that encapsulation is mediated by binding of the PduP targeting sequence to a short C-terminal helix of the PduA shell protein. In vitro studies indicated binding between PduP and PduA (and PduJ) but not other MCP shell proteins. Alanine scanning mutagenesis determined that the key residues involved in binding are E7, I10, and L14 of PduP and H81, V84, and L88 of PduA. In vivo targeting studies indicated that the binding between the N terminus of PduP and the C terminus of PduA is critical for encapsulation of PduP within the Pdu MCP. Structural models suggest that the N terminus of PduP and C terminus of PduA both form helical structures that bind one another via the key residues identified by mutagenesis. Cumulatively, these results show that the N-terminal targeting sequence of PduP promotes its encapsulation by binding to MCP shell proteins. This is a unique report determining the mechanism by which a MCP targeting sequence functions. We propose that specific interactions between the termini of shell proteins and lumen enzymes have general importance for guiding the assembly and the higher level organization of bacterial MCPs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. - PubMed
-
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. - PubMed
-
- Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): Functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–1461. - PubMed
-
- Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

