Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution
- PMID: 22927429
- PMCID: PMC3443142
- DOI: 10.1073/pnas.1207846109
Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution
Abstract
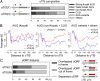
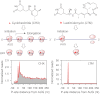
Understanding translational control in gene expression relies on precise and comprehensive determination of translation initiation sites (TIS) across the entire transcriptome. The recently developed ribosome-profiling technique enables global translation analysis, providing a wealth of information about both the position and the density of ribosomes on mRNAs. Here we present an approach, global translation initiation sequencing, applying in parallel the ribosome E-site translation inhibitors lactimidomycin and cycloheximide to achieve simultaneous detection of both initiation and elongation events on a genome-wide scale. This approach provides a view of alternative translation initiation in mammalian cells with single-nucleotide resolution. Systemic analysis of TIS positions supports the ribosome linear-scanning mechanism in TIS selection. The alternative TIS positions and the associated ORFs identified by global translation initiation sequencing are conserved between human and mouse cells, implying physiological significance of alternative translation. Our study establishes a practical platform for uncovering the hidden coding potential of the transcriptome and offers a greater understanding of the complexity of translation initiation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

