A viral, transporter associated with antigen processing (TAP)-independent, high affinity ligand with alternative interactions endogenously presented by the nonclassical human leukocyte antigen E class I molecule
- PMID: 22927436
- PMCID: PMC3471699
- DOI: 10.1074/jbc.M112.362293
A viral, transporter associated with antigen processing (TAP)-independent, high affinity ligand with alternative interactions endogenously presented by the nonclassical human leukocyte antigen E class I molecule
Abstract
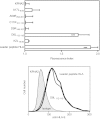
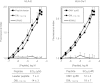
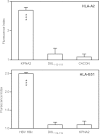
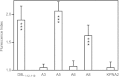
The transporter associated with antigen processing (TAP) enables the flow of viral peptides generated in the cytosol by the proteasome and other proteases to the endoplasmic reticulum, where they complex with nascent human leukocyte antigen (HLA) class I. Later, these peptide-HLA class I complexes can be recognized by CD8(+) lymphocytes. Cancerous cells and infected cells in which TAP is blocked, as well as individuals with unusable TAP complexes, are able to present peptides on HLA class I by generating them through TAP-independent processing pathways. Here, we identify a physiologically processed HLA-E ligand derived from the D8L protein in TAP-deficient vaccinia virus-infected cells. This natural high affinity HLA-E class I ligand uses alternative interactions to the anchor motifs previously described to be presented on nonclassical HLA class I molecules. This octameric peptide was also presented on HLA-Cw1 with similar binding affinity on both classical and nonclassical class I molecules. In addition, this viral peptide inhibits HLA-E-mediated cytolysis by natural killer cells. Comparison between the amino acid sequences of the presenting HLA-E and HLA-Cw1 alleles revealed a shared structural motif in both HLA class molecules, which could be related to their observed similar cross-reactivity affinities. This motif consists of several residues located on the floor of the peptide-binding site. These data expand the role of HLA-E as an antigen-presenting molecule.
Figures



References
-
- Kloetzel P. M., Ossendorp F. (2004) Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 16, 76–81 - PubMed
-
- Cerundolo V., de la Salle H. (2006) Description of HLA class I- and CD8-deficient patients. Insights into the function of cytotoxic T lymphocytes and NK cells in host defense. Semin. Immunol. 18, 330–336 - PubMed
-
- Van Kaer L., Ashton-Rickardt P. G., Ploegh H. L., Tonegawa S. (1992) TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4–8+ T cells. Cell 71, 1205–1214 - PubMed
-
- Voldby Larsen M., Nielsen M., Weinzierl A., Lund O. (2006) TAP-independent MHC class I presentation. Curr. Immunol. Rev. 2, 233–245
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

