Artificial neural networks trained to detect viral and phage structural proteins
- PMID: 22927809
- PMCID: PMC3426561
- DOI: 10.1371/journal.pcbi.1002657
Artificial neural networks trained to detect viral and phage structural proteins
Abstract
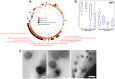
Phages play critical roles in the survival and pathogenicity of their hosts, via lysogenic conversion factors, and in nutrient redistribution, via cell lysis. Analyses of phage- and viral-encoded genes in environmental samples provide insights into the physiological impact of viruses on microbial communities and human health. However, phage ORFs are extremely diverse of which over 70% of them are dissimilar to any genes with annotated functions in GenBank. Better identification of viruses would also aid in better detection and diagnosis of disease, in vaccine development, and generally in better understanding the physiological potential of any environment. In contrast to enzymes, viral structural protein function can be much more challenging to detect from sequence data because of low sequence conservation, few known conserved catalytic sites or sequence domains, and relatively limited experimental data. We have designed a method of predicting phage structural protein sequences that uses Artificial Neural Networks (ANNs). First, we trained ANNs to classify viral structural proteins using amino acid frequency; these correctly classify a large fraction of test cases with a high degree of specificity and sensitivity. Subsequently, we added estimates of protein isoelectric points as a feature to ANNs that classify specialized families of proteins, namely major capsid and tail proteins. As expected, these more specialized ANNs are more accurate than the structural ANNs. To experimentally validate the ANN predictions, several ORFs with no significant similarities to known sequences that are ANN-predicted structural proteins were examined by transmission electron microscopy. Some of these self-assembled into structures strongly resembling virion structures. Thus, our ANNs are new tools for identifying phage and potential prophage structural proteins that are difficult or impossible to detect by other bioinformatic analysis. The networks will be valuable when sequence is available but in vitro propagation of the phage may not be practical or possible.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Functional and comparative genome analysis of novel virulent actinophages belonging to Streptomyces flavovirens.BMC Microbiol. 2017 Mar 3;17(1):51. doi: 10.1186/s12866-017-0940-7. BMC Microbiol. 2017. PMID: 28257628 Free PMC article.
-
Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54.J Mol Biol. 2003 Aug 15;331(3):605-22. doi: 10.1016/s0022-2836(03)00763-0. J Mol Biol. 2003. PMID: 12899832
-
Genome organization and characterization of the virulent lactococcal phage 1358 and its similarities to Listeria phages.Appl Environ Microbiol. 2010 Mar;76(5):1623-32. doi: 10.1128/AEM.02173-09. Epub 2010 Jan 8. Appl Environ Microbiol. 2010. PMID: 20061452 Free PMC article.
-
Metaviromics coupled with phage-host identification to open the viral 'black box'.J Microbiol. 2021 Mar;59(3):311-323. doi: 10.1007/s12275-021-1016-9. Epub 2021 Feb 23. J Microbiol. 2021. PMID: 33624268 Review.
-
Artificial neural networks in contemporary toxicology research.Chem Biol Interact. 2023 Jan 5;369:110269. doi: 10.1016/j.cbi.2022.110269. Epub 2022 Nov 17. Chem Biol Interact. 2023. PMID: 36402212 Review.
Cited by
-
Identification of Phage Viral Proteins With Hybrid Sequence Features.Front Microbiol. 2019 Mar 26;10:507. doi: 10.3389/fmicb.2019.00507. eCollection 2019. Front Microbiol. 2019. PMID: 30972038 Free PMC article.
-
Identification of Known and Novel Recurrent Viral Sequences in Data from Multiple Patients and Multiple Cancers.Viruses. 2016 Feb 19;8(2):53. doi: 10.3390/v8020053. Viruses. 2016. PMID: 26907326 Free PMC article.
-
The human gut virome: a multifaceted majority.Front Microbiol. 2015 Sep 11;6:918. doi: 10.3389/fmicb.2015.00918. eCollection 2015. Front Microbiol. 2015. PMID: 26441861 Free PMC article. Review.
-
PVPred-SCM: Improved Prediction and Analysis of Phage Virion Proteins Using a Scoring Card Method.Cells. 2020 Feb 3;9(2):353. doi: 10.3390/cells9020353. Cells. 2020. PMID: 32028709 Free PMC article.
-
Predicting Bacteriophage Enzymes and Hydrolases by Using Combined Features.Front Bioeng Biotechnol. 2020 Mar 24;8:183. doi: 10.3389/fbioe.2020.00183. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32266225 Free PMC article.
References
-
- Rohwer F, Prangishvili D, Lindell D (2009) Roles of viruses in the environment. Environ Microbiol 11: 2771–2774. - PubMed
-
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, et al. (2008) Functional metagenomic profiling of nine biomes. Nature 452: 629–632. - PubMed
-
- Suttle CA (2007) Marine viruses-major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

