Cytochrome P450 2B diversity and dietary novelty in the herbivorous, desert woodrat (Neotoma lepida)
- PMID: 22927909
- PMCID: PMC3425548
- DOI: 10.1371/journal.pone.0041510
Cytochrome P450 2B diversity and dietary novelty in the herbivorous, desert woodrat (Neotoma lepida)
Abstract
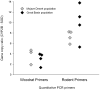
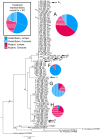
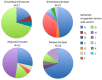
Detoxification enzymes play a key role in plant-herbivore interactions, contributing to the on-going evolution of ecosystem functional diversity. Mammalian detoxification systems have been well studied by the medical and pharmacological industries to understand human drug metabolism; however, little is known of the mechanisms employed by wild herbivores to metabolize toxic plant secondary compounds. Using a wild rodent herbivore, the desert woodrat (Neotoma lepida), we investigated genomic structural variation, sequence variability, and expression patterns in a multigene subfamily involved in xenobiotic metabolism, cytochrome P450 2B (CYP2B). We hypothesized that differences in CYP2B expression and sequence diversity could explain differential abilities of woodrat populations to consume native plant toxins. Woodrats from two distinct populations were fed diets supplemented with either juniper (Juniperus osteosperma) or creosote bush (Larrea tridentata), plants consumed by woodrats in their respective desert habitats. We used Southern blot and quantitative PCR to determine that the genomic copy number of CYP2B in both populations was equivalent, and similar in number to known rodent copy number. We compared CYP2B expression patterns and sequence diversity using cloned hepatic CYP2B cDNA. The resulting sequences were very diverse, and clustered into four major clades by amino acid similarity. Sequences from the experimental treatments were distributed non-randomly across a CYP2B tree, indicating unique expression patterns from woodrats on different diets and from different habitats. Furthermore, within each major CYP2B clade, sequences shared a unique combination of amino acid residues at 13 sites throughout the protein known to be important for CYP2B enzyme function, implying differences in the function of each major CYP2B variant. This work is the most comprehensive investigation of the genetic diversity of a detoxification enzyme subfamily in a wild mammalian herbivore, and contributes an initial genetic framework to our understanding of how a wild herbivore responds to critical changes in its diet.
Conflict of interest statement
Figures
Similar articles
-
Functional characterization of cytochromes P450 2B from the desert woodrat Neotoma lepida.Toxicol Appl Pharmacol. 2014 Feb 1;274(3):393-401. doi: 10.1016/j.taap.2013.12.005. Epub 2013 Dec 19. Toxicol Appl Pharmacol. 2014. PMID: 24361551 Free PMC article.
-
Role of cytochrome P450 2B sequence variation and gene copy number in facilitating dietary specialization in mammalian herbivores.Mol Ecol. 2018 Feb;27(3):723-736. doi: 10.1111/mec.14480. Epub 2018 Jan 29. Mol Ecol. 2018. PMID: 29319892
-
Turning Over an Old Leaf- do Mammalian Herbivores Retain the Ability to Biotransform Toxic Ancestral Diets?J Chem Ecol. 2025 Apr 15;51(2):48. doi: 10.1007/s10886-025-01599-x. J Chem Ecol. 2025. PMID: 40234269 Free PMC article.
-
The Woodrat Gut Microbiota as an Experimental System for Understanding Microbial Metabolism of Dietary Toxins.Front Microbiol. 2016 Jul 28;7:1165. doi: 10.3389/fmicb.2016.01165. eCollection 2016. Front Microbiol. 2016. PMID: 27516760 Free PMC article. Review.
-
Evolution of the cytochrome P450 superfamily: sequence alignments and pharmacogenetics.Mutat Res. 1998 Jun;410(3):245-70. doi: 10.1016/s1383-5742(97)00040-9. Mutat Res. 1998. PMID: 9630657 Review.
Cited by
-
Human cytochrome P450 2B6 genetic variability in Botswana: a case of haplotype diversity and convergent phenotypes.Sci Rep. 2018 Mar 20;8(1):4912. doi: 10.1038/s41598-018-23350-1. Sci Rep. 2018. PMID: 29559695 Free PMC article.
-
Toxin tolerance across landscapes: Ecological exposure not a prerequisite.Funct Ecol. 2022 Aug;36(8):2119-2131. doi: 10.1111/1365-2435.14093. Epub 2022 May 21. Funct Ecol. 2022. PMID: 37727272 Free PMC article.
-
Functional characterization of cytochromes P450 2B from the desert woodrat Neotoma lepida.Toxicol Appl Pharmacol. 2014 Feb 1;274(3):393-401. doi: 10.1016/j.taap.2013.12.005. Epub 2013 Dec 19. Toxicol Appl Pharmacol. 2014. PMID: 24361551 Free PMC article.
-
Structure-Function Analysis of Mammalian CYP2B Enzymes Using 7-Substituted Coumarin Derivatives as Probes: Utility of Crystal Structures and Molecular Modeling in Understanding Xenobiotic Metabolism.Mol Pharmacol. 2016 Apr;89(4):435-45. doi: 10.1124/mol.115.102111. Epub 2016 Jan 29. Mol Pharmacol. 2016. PMID: 26826176 Free PMC article.
-
Warmer ambient temperatures depress liver function in a mammalian herbivore.Biol Lett. 2013 Oct 23;9(5):20130562. doi: 10.1098/rsbl.2013.0562. Biol Lett. 2013. PMID: 24046878 Free PMC article.
References
-
- Stamp N (2003) Out of the quagmire of plant defense hypotheses. Q Rev Biol 78: 23–55. - PubMed
-
- Klaassen C (2001) Cararett and Doull's Toxicology: The Basic Science of Poisons. New York: McGraw Hill.
-
- Ma Q, Lu AY (2008) The challenges of dealing with promiscuous drug-metabolizing enzymes, receptors and transporters. Curr Drug Metab 9: 374–383. - PubMed
-
- Baker ME (2005) Xenobiotics and the evolution of multicellular animals: Emergence and diversification of ligand-activated transcription factors'. Integrative and Comparative Biology 45: 172–178. - PubMed
-
- Gonzalez-Duarte R, Albalat R (2005) Merging protein, gene and genomic data: the evolution of the MDR-ADH family. Heredity 95: 184–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

