Ultrasensitive cDNA detection of dengue virus RNA using electrochemical nanoporous membrane-based biosensor
- PMID: 22927927
- PMCID: PMC3426509
- DOI: 10.1371/journal.pone.0042346
Ultrasensitive cDNA detection of dengue virus RNA using electrochemical nanoporous membrane-based biosensor
Abstract
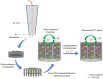
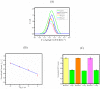
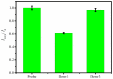
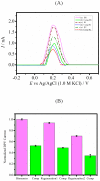
A nanoporous alumina membrane-based ultrasensitive DNA biosensor is constructed using 5'-aminated DNA probes immobilized onto the alumina channel walls. Alumina nanoporous membrane-like structure is carved over platinum wire electrode of 76 µm diameter dimension by electrochemical anodization. The hybridization of complementary target DNA with probe DNA molecules attached inside the pores influences the pore size and ionic conductivity. The biosensor demonstrates linear range over 6 order of magnitude with ultrasensitive detection limit of 9.55×10(-12) M for the quantification of ss-31 mer DNA sequence. Its applicability is challenged against real time cDNA PCR sample of dengue virus serotype1 derived from asymmetric PCR. Excellent specificity down to one nucleotide mismatch in target DNA sample of DENV3 is also demonstrated.
Conflict of interest statement
Figures
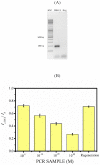
References
-
- Lee KS, Lo S, Tan SSY, Chua R, Tan LK, et al. (2012) Dengue virus surveillance in Singapore reveals high viral diversity through multiple introductions and in situ evolution. Infection, Genetics and Evolution 12: 77–85. - PubMed
-
- Mackay IM (2004) Real-time PCR in the microbiology laboratory. Clinical Microbiology and Infection 10: 190–212. - PubMed
-
- Kang M, Trofin L, Mota MO, Martin CR (2005) Protein capture in silica nanotube membrane 3-D microwell arrays. Anal Chem 77: 6243–6249. - PubMed
-
- Cheng MS, Lau SH, Chow VT, Toh CS (2011) Membrane-based electrochemical nanobiosensor for Escherichia coli detection and analysis of cells viability. Environ Sci Technol 45: 6453–6459. - PubMed
-
- Nguyen BTT, Koh G, Lim HS, Chua AJS, Ng MML, et al. (2009) Membrane-Based Electrochemical Nanobiosensor for the Detection of Virus. Anal Chem 81: 7226–7234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

