Developmental dynamics of post-selection thymic DN iNKT
- PMID: 22927977
- PMCID: PMC3425480
- DOI: 10.1371/journal.pone.0043509
Developmental dynamics of post-selection thymic DN iNKT
Abstract
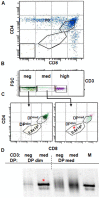
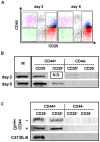
Background: Invariant natural killer T (iNKT) cells develop in the thymus and branch off from the maturation pathway of conventional T cell at the DP stage. While different stages of iNKT cellular development have been defined, the actual time that iNKT cell precursors spend at each stage is still unknown.
Methodology/principal finding: Here we report on maturation dynamics of post-selection DN iNKT cells by injecting wild-type DP(dim) thymocytes into the thymus of TCRα(-/-) mice and using the Vα14-Jα18 rearrangements as a molecular marker to follow the maturation dynamics of these cells.
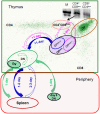
Conclusion/significance: This study shows that the developmental dynamics of DN iNKT cells in DP(dim) are very rapid and that it takes less than 1 day to down-regulate CD4 and CD8 and become DN. These DN cells are precursors of peripheral DN iNKT cells and appear in the spleen in 1-2 days. Thymic DN iNKT residents are predominantly derived from cells that quickly return from the periphery. The expansion of a very small subset of DN iNKT precursors could also play a small role in this process. These data are an example of measuring T cell maturation in the thymus and show that the maturation dynamics of selected DN iNKT cells fall within the same general time frame as conventional αβ T cells.
Conflict of interest statement
Figures



Similar articles
-
Murine CD8+ Invariant Natural Killer T Cells are Negatively Selected by CD1d Expressed on Thymic Epithelial Cells and Dendritic Cells.Immunol Invest. 2018 Jan;47(1):89-100. doi: 10.1080/08820139.2017.1385621. Epub 2017 Nov 3. Immunol Invest. 2018. PMID: 29099628
-
Identification of CD4(-)CD8(-) double-negative natural killer T cell precursors in the thymus.PLoS One. 2008;3(11):e3688. doi: 10.1371/journal.pone.0003688. Epub 2008 Nov 10. PLoS One. 2008. PMID: 18997862 Free PMC article.
-
Distinct and overlapping effector functions of expanded human CD4+, CD8α+ and CD4-CD8α- invariant natural killer T cells.PLoS One. 2011;6(12):e28648. doi: 10.1371/journal.pone.0028648. Epub 2011 Dec 12. PLoS One. 2011. PMID: 22174854 Free PMC article.
-
Thymus medulla fosters generation of natural Treg cells, invariant γδ T cells, and invariant NKT cells: what we learn from intrathymic migration.Eur J Immunol. 2015 Mar;45(3):652-60. doi: 10.1002/eji.201445108. Epub 2015 Feb 13. Eur J Immunol. 2015. PMID: 25615828 Free PMC article. Review.
-
New Genetically Manipulated Mice Provide Insights Into the Development and Physiological Functions of Invariant Natural Killer T Cells.Front Immunol. 2018 Jun 14;9:1294. doi: 10.3389/fimmu.2018.01294. eCollection 2018. Front Immunol. 2018. PMID: 29963043 Free PMC article. Review.
Cited by
-
Protective effects of Ganoderma lucidum spores on estradiol benzoate-induced TEC apoptosis and compromised double-positive thymocyte development.Front Pharmacol. 2024 Aug 16;15:1419881. doi: 10.3389/fphar.2024.1419881. eCollection 2024. Front Pharmacol. 2024. PMID: 39221140 Free PMC article.
References
-
- Gapin L, Matsuda JL, Surh CD, Kronenberg M (2001) NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol 2: 971–978. - PubMed
-
- Dao T, Guo D, Ploss A, Stolzer A, Saylor C, et al. (2004) Development of CD1d-restricted NKT cells in the mouse thymus. Eur J Immunol 34: 3542–3552. - PubMed
-
- Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, et al. (2005) Genetic evidence supporting selection of the Vα14 iNKT cell lineage from double-positive thymocyte precursors. Immunity 22: 705–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

