Pathogen webs in collapsing honey bee colonies
- PMID: 22927991
- PMCID: PMC3424165
- DOI: 10.1371/journal.pone.0043562
Pathogen webs in collapsing honey bee colonies
Abstract
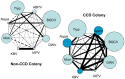
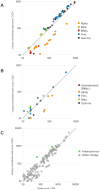
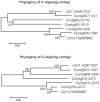
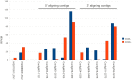
Recent losses in honey bee colonies are unusual in their severity, geographical distribution, and, in some cases, failure to present recognized characteristics of known disease. Domesticated honey bees face numerous pests and pathogens, tempting hypotheses that colony collapses arise from exposure to new or resurgent pathogens. Here we explore the incidence and abundance of currently known honey bee pathogens in colonies suffering from Colony Collapse Disorder (CCD), otherwise weak colonies, and strong colonies from across the United States. Although pathogen identities differed between the eastern and western United States, there was a greater incidence and abundance of pathogens in CCD colonies. Pathogen loads were highly covariant in CCD but not control hives, suggesting that CCD colonies rapidly become susceptible to a diverse set of pathogens, or that co-infections can act synergistically to produce the rapid depletion of workers that characterizes the disorder. We also tested workers from a CCD-free apiary to confirm that significant positive correlations among pathogen loads can develop at the level of individual bees and not merely as a secondary effect of CCD. This observation and other recent data highlight pathogen interactions as important components of bee disease. Finally, we used deep RNA sequencing to further characterize microbial diversity in CCD and non-CCD hives. We identified novel strains of the recently described Lake Sinai viruses (LSV) and found evidence of a shift in gut bacterial composition that may be a biomarker of CCD. The results are discussed with respect to host-parasite interactions and other environmental stressors of honey bees.
Conflict of interest statement
Figures
References
-
- Gallai N, Salles JM, Settele J, Vaissière BE (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecological Economics 68: 810–821.
-
- Aizen MA, Harder LD (2009) The global stock of domesticated honey bees is growing slower than agricultural demand for pollination. Current Biology 19: 915–918. - PubMed
-
- Potts SG, Roberts SPM, Dean R, Marris G, Brown MA, et al. (2010) Declines of managed honey bees and beekeepers in Europe. Journal of Apicultural Research 49: 15–22.
-
- VanEngelsdorp D, Hayes Jr J, Underwood RM, Pettis JS (2011) A survey of managed honey bee colony losses in the United States, fall 2009 to winter 2010. Journal of Apicultural Research 50: 1–10.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

