Context and force field dependence of the loss of protein backbone entropy upon folding using realistic denatured and native state ensembles
- PMID: 22928488
- PMCID: PMC3464005
- DOI: 10.1021/ja3064028
Context and force field dependence of the loss of protein backbone entropy upon folding using realistic denatured and native state ensembles
Abstract
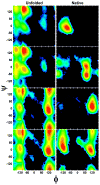
The loss of conformational entropy is the largest unfavorable quantity affecting a protein's stability. We calculate the reduction in the number of backbone conformations upon folding using the distribution of backbone dihedral angles (ϕ,ψ) obtained from an experimentally validated denatured state model, along with all-atom simulations for both the denatured and native states. The average loss of entropy per residue is TΔS(BB)(U-N) = 0.7, 0.9, or 1.1 kcal·mol(-1) at T = 298 K, depending on the force field used, with a 0.6 kcal·mol(-1) dispersion across the sequence. The average equates to a decrease of a factor of 3-7 in the number of conformations available per residue (f = Ω(Denatured)/Ω(Native)) or to a total of f(tot) = 3(n)-7(n) for an n residue protein. Our value is smaller than most previous estimates where f = 7-20, that is, our computed TΔS(BB)(U-N) is smaller by 10-100 kcal mol(-1) for n = 100. The differences emerge from our use of realistic native and denatured state ensembles as well as from the inclusion of accurate local sequence preferences, neighbor effects, and correlated motions (vibrations), in contrast to some previous studies that invoke gross assumptions about the entropy in either or both states. We find that the loss of entropy primarily depends on the local environment and less on properties of the native state, with the exception of α-helical residues in some force fields.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Stites WE, Pranata J. Proteins. 1995;22:132–40. - PubMed
-
- Meirovitch H. Curr Opin Struct Biol. 2007;17:181–6. - PubMed
-
- D’Aquino JA, Gomez J, Hilser VJ, Lee KH, Amzel LM, Freire E. Proteins. 1996;25:143–56. - PubMed
-
- Yang D, Kay LE. J Mol Biol. 1996;263:369–82. - PubMed
-
- Zaman MH, Shen MY, Berry RS, Freed KF, Sosnick TR. J Mol Biol. 2003;331:693–711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

