Comparative chemogenomics to examine the mechanism of action of dna-targeted platinum-acridine anticancer agents
- PMID: 22928710
- PMCID: PMC3500413
- DOI: 10.1021/cb300320d
Comparative chemogenomics to examine the mechanism of action of dna-targeted platinum-acridine anticancer agents
Abstract
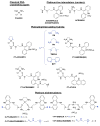
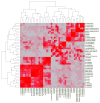
Platinum-based drugs have been used to successfully treat diverse cancers for several decades. Cisplatin, the original compound of this class, cross-links DNA, resulting in cell cycle arrest and cell death via apoptosis. Cisplatin is effective against several tumor types, yet it exhibits toxic side effects and tumors often develop resistance. To mitigate these liabilities while maintaining potency, we generated a library of non-classical platinum-acridine hybrid agents and assessed their mechanisms of action using a validated genome-wide screening approach in Saccharomyces cerevisiae and in the distantly related yeast Schizosaccharomyces pombe. Chemogenomic profiles from both S. cerevisiae and S. pombe demonstrate that several of the platinum-acridines damage DNA differently than cisplatin based on their requirement for distinct modules of DNA repair.
Figures



Similar articles
-
Redesigning the DNA-targeted chromophore in platinum-acridine anticancer agents: a structure-activity relationship study.Chemistry. 2014 Dec 1;20(49):16174-87. doi: 10.1002/chem.201404845. Epub 2014 Oct 10. Chemistry. 2014. PMID: 25302716 Free PMC article.
-
Design, synthesis, and biological activity of a novel non-cisplatin-type platinum-acridine pharmacophore.J Med Chem. 2001 Dec 6;44(25):4492-6. doi: 10.1021/jm010293m. J Med Chem. 2001. PMID: 11728195
-
Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C.Cancer Res. 2004 Jun 1;64(11):3940-8. doi: 10.1158/0008-5472.CAN-03-3113. Cancer Res. 2004. PMID: 15173006
-
Cellular processing of platinum anticancer drugs.Nat Rev Drug Discov. 2005 Apr;4(4):307-20. doi: 10.1038/nrd1691. Nat Rev Drug Discov. 2005. PMID: 15789122 Review.
-
Adenine-N3 in the DNA minor groove - an emerging target for platinum containing anticancer pharmacophores.Anticancer Agents Med Chem. 2007 Jan;7(1):125-38. doi: 10.2174/187152007779313991. Anticancer Agents Med Chem. 2007. PMID: 17266509 Review.
Cited by
-
Using fluorescent post-labeling to probe the subcellular localization of DNA-targeted platinum anticancer agents.Angew Chem Int Ed Engl. 2013 Mar 18;52(12):3350-4. doi: 10.1002/anie.201210079. Epub 2013 Feb 20. Angew Chem Int Ed Engl. 2013. PMID: 23427109 Free PMC article. No abstract available.
-
Acridine as an Anti-Tumour Agent: A Critical Review.Molecules. 2022 Dec 26;28(1):193. doi: 10.3390/molecules28010193. Molecules. 2022. PMID: 36615391 Free PMC article. Review.
-
Using phosphine ligands with a biological role to modulate reactivity in novel platinum complexes.R Soc Open Sci. 2018 Feb 21;5(2):171340. doi: 10.1098/rsos.171340. eCollection 2018 Feb. R Soc Open Sci. 2018. PMID: 29515851 Free PMC article.
-
From beer to breadboards: yeast as a force for biological innovation.Genome Biol. 2024 Jan 4;25(1):10. doi: 10.1186/s13059-023-03156-9. Genome Biol. 2024. PMID: 38178179 Free PMC article. Review.
-
Redesigning the DNA-targeted chromophore in platinum-acridine anticancer agents: a structure-activity relationship study.Chemistry. 2014 Dec 1;20(49):16174-87. doi: 10.1002/chem.201404845. Epub 2014 Oct 10. Chemistry. 2014. PMID: 25302716 Free PMC article.
References
-
- Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. - PubMed
-
- Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res. 2001;478:23–43. - PubMed
-
- Wong E, Giandomenico CM. Current status of platinum-based antitumor drugs. Chemical reviews. 1999;99:2451–2466. - PubMed
-
- Cleare MJ, Hoeschele JD. Studies on the antitumor activity of group VIII transition metal complexes. Part I. Platinum (II) complexes. Bioinorganic Chemistry. 1973;2:187–210.
-
- Manzotti C, Pratesi G, Menta E, Di Domenico R, Cavalletti E, Fiebig HH, Kelland LR, Farrell N, Polizzi D, Supino R, Pezzoni G, Zunino F. BBR 3464: a novel triplatinum complex, exhibiting a preclinical profile of antitumor efficacy different from cisplatin. Clin Cancer Res. 2000;6:2626–2634. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

