Regulation of multiple tip formation by caffeine in cellular slime molds
- PMID: 22928977
- PMCID: PMC3488011
- DOI: 10.1186/1471-213X-12-26
Regulation of multiple tip formation by caffeine in cellular slime molds
Abstract
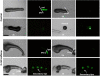
Background: The multicellular slug in Dictyostelium has a single tip that acts as an organising centre patterning the rest of the slug. High adenosine levels at the tip are believed to be responsible for this tip dominance and the adenosine antagonist, caffeine overrides this dominance promoting multiple tip formation.
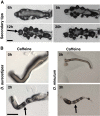
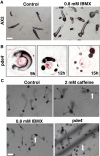
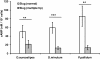
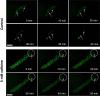
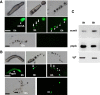
Results: Caffeine induced multiple tip effect is conserved in all the Dictyostelids tested. Two key components of cAMP relay namely, cAMP phosphodiesterase (Pde4) and adenyl cyclase-A (AcaA) levels get reduced during secondary tip formation in Dictyostelium discoideum. Pharmacological inhibition of cAMP phosphodiesterase also resulted in multiple tips. Caffeine reduces cAMP levels by 16.4, 2.34, 4.71 and 6.30 folds, respectively in D. discoideum, D. aureostipes, D. minutum and Polysphondylium pallidum. We propose that altered cAMP levels, perturbed cAMP gradient and impaired signalling may be the critical factors for the origin of multiple tips in other Dictyostelids as well. In the presence of caffeine, slug cell movement gets impaired and restricted. The cell type specific markers, ecmA (prestalk) and pspA (prespore) cells are not equally contributing during additional tip formation. During additional tip emergence, prespore cells transdifferentiate to compensate the loss of prestalk cells.
Conclusion: Caffeine decreases adenyl cyclase-A (AcaA) levels and as a consequence low cAMP is synthesised altering the gradient. Further if cAMP phosphodiesterase (Pde4) levels go down in the presence of caffeine, the cAMP gradient breaks down. When there is no cAMP gradient, directional movement is inhibited and might favour re-differentiation of prespore to prestalk cells.
Figures



Similar articles
-
Regulation of aggregate size and pattern by adenosine and caffeine in cellular slime molds.BMC Dev Biol. 2012 Jan 23;12:5. doi: 10.1186/1471-213X-12-5. BMC Dev Biol. 2012. PMID: 22269093 Free PMC article.
-
Influence of cyclic AMP and hydrolysis products on cell type regulation in Dictyostelium discoideum.J Embryol Exp Morphol. 1985 Apr;86:19-37. J Embryol Exp Morphol. 1985. PMID: 2993468
-
Analysis of cell movement and signalling during ring formation in an activated G alpha1 mutant of Dictyostelium discoideum that is defective in prestalk zone formation.Dev Biol. 1997 Jan 1;181(1):79-90. doi: 10.1006/dbio.1996.8447. Dev Biol. 1997. PMID: 9015266
-
The prestalk-prespore pattern in cellular slime molds.Differentiation. 1979;14(1-2):1-22. doi: 10.1111/j.1432-0436.1979.tb01006.x. Differentiation. 1979. PMID: 225235 Review.
-
Regulation of Dictyostelium morphogenesis by cAMP-dependent protein kinase.Philos Trans R Soc Lond B Biol Sci. 1993 Jun 29;340(1293):305-13. doi: 10.1098/rstb.1993.0072. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8103933 Review.
Cited by
-
Diverse Roles of the Multiple Phosphodiesterases in the Regulation of Cyclic Nucleotide Signaling in Dictyostelium.Cells. 2025 Apr 1;14(7):522. doi: 10.3390/cells14070522. Cells. 2025. PMID: 40214475 Free PMC article. Review.
-
An integrated, cross-regulation pathway model involving activating/adaptive and feed-forward/feed-back loops for directed oscillatory cAMP signal-relay/response during the development of Dictyostelium.Front Cell Dev Biol. 2024 Jan 31;11:1263316. doi: 10.3389/fcell.2023.1263316. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38357530 Free PMC article.
References
-
- Bonner JT. The cellular slime molds. Second edition, Princeton University Press; 1967.
-
- Schindler J, Sussman M. Inhibition by ammonia of intracellular cAMP accumulation in Dictyostelium discoideum: Its significance for the regulation of morphogenesis. Dev. Genetics. 1967;1(1):13–20.
-
- Kay RR, Berks M, Traynor D. Morphogen hunting in Dictyostelium. Development Supplement. 1989;107:81–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

