Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury
- PMID: 22929533
- PMCID: PMC3511201
- DOI: 10.1186/1479-5876-10-174
Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury
Abstract
Background: Clinical dogma is that healthy urine is sterile and the presence of bacteria with an inflammatory response is indicative of urinary tract infection (UTI). Asymptomatic bacteriuria (ABU) represents the state in which bacteria are present but the inflammatory response is negligible. Differentiating ABU from UTI is diagnostically challenging, but critical because overtreatment of ABU can perpetuate antimicrobial resistance while undertreatment of UTI can result in increased morbidity and mortality. In this study, we describe key characteristics of the healthy and ABU urine microbiomes utilizing 16S rRNA gene (16S rDNA) sequencing and metaproteomics, with the future goal of utilizing this information to personalize the treatment of UTI based on key individual characteristics.
Methods: A cross-sectional study of 26 healthy controls and 27 healthy subjects at risk for ABU due to spinal cord injury-related neuropathic bladder (NB) was conducted. Of the 27 subjects with NB, 8 voided normally, 8 utilized intermittent catheterization, and 11 utilized indwelling Foley urethral catheterization for bladder drainage. Urine was obtained by clean catch in voiders, or directly from the catheter in subjects utilizing catheters. Urinalysis, urine culture and 16S rDNA sequencing were performed on all samples, with metaproteomic analysis performed on a subsample.
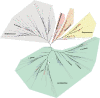
Results: A total of 589454 quality-filtered 16S rDNA sequence reads were processed through a NextGen 16S rDNA analysis pipeline. Urine microbiomes differ by normal bladder function vs. NB, gender, type of bladder catheter utilized, and duration of NB. The top ten bacterial taxa showing the most relative abundance and change among samples were Lactobacillales, Enterobacteriales, Actinomycetales, Bacillales, Clostridiales, Bacteroidales, Burkholderiales, Pseudomonadales, Bifidobacteriales and Coriobacteriales. Metaproteomics confirmed the 16S rDNA results, and functional human protein-pathogen interactions were noted in subjects where host defenses were initiated.
Conclusions: Counter to clinical belief, healthy urine is not sterile. The healthy urine microbiome is characterized by a preponderance of Lactobacillales in women and Corynebacterium in men. The presence and duration of NB and method of urinary catheterization alter the healthy urine microbiome. An integrated approach of 16S rDNA sequencing with metaproteomics improves our understanding of healthy urine and facilitates a more personalized approach to prevention and treatment of infection.
Figures




References
-
- Infectious disease, chapter 7, urinary tract infections. http://pathmicro.med.sc.edu/infectiousdisease/UrinaryTractInfections.htm.
-
- HHS action plan to prevent healthcare-sssociated infections. http://www.hhs.gov/ash/initiatives/hai/actionplan.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

