Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit
- PMID: 22929616
- PMCID: PMC3435506
- DOI: 10.1038/msb.2012.39
Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit
Abstract
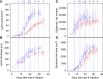
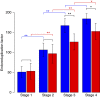
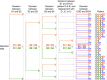
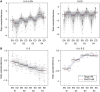
Leaves have a central role in plant energy capture and carbon conversion and therefore must continuously adapt their development to prevailing environmental conditions. To reveal the dynamic systems behaviour of leaf development, we profiled Arabidopsis leaf number six in depth at four different growth stages, at both the end-of-day and end-of-night, in plants growing in two controlled experimental conditions: short-day conditions with optimal soil water content and constant reduced soil water conditions. We found that the lower soil water potential led to reduced, but prolonged, growth and an adaptation at the molecular level without a drought stress response. Clustering of the protein and transcript data using a decision tree revealed different patterns in abundance changes across the growth stages and between end-of-day and end-of-night that are linked to specific biological functions. Correlations between protein and transcript levels depend on the time-of-day and also on protein localisation and function. Surprisingly, only very few of >1700 quantified proteins showed diurnal abundance fluctuations, despite strong fluctuations at the transcript level.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Achard P, Cheng H, Grauwe LD, Decat J, Schoutteten H, Moritz T, Straeten DVD, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 - PubMed
-
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29: 2216–2227 - PubMed
-
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
-
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 - PubMed
-
- Anastasiou E, Lenhard M (2007) Growing up to one’s standard. Curr Opin Plant Biol 10: 63–69 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

