Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model
- PMID: 22930817
- PMCID: PMC3443116
- DOI: 10.1073/pnas.1211733109
Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model
Erratum in
-
Correction for Song et al., Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model.Proc Natl Acad Sci U S A. 2015 Nov 3;112(44):E6079. doi: 10.1073/pnas.1518753112. Epub 2015 Oct 26. Proc Natl Acad Sci U S A. 2015. PMID: 26504207 Free PMC article. No abstract available.
Abstract
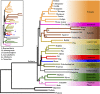
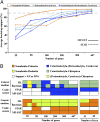
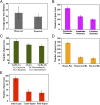
The reconstruction of the Tree of Life has relied almost entirely on concatenation methods, which do not accommodate gene tree heterogeneity, a property that simulations and theory have identified as a likely cause of incongruent phylogenies. However, this incongruence has not yet been demonstrated in empirical studies. Several key relationships among eutherian mammals remain controversial and conflicting among previous studies, including the root of eutherian tree and the relationships within Euarchontoglires and Laurasiatheria. Both bayesian and maximum-likelihood analysis of genome-wide data of 447 nuclear genes from 37 species show that concatenation methods indeed yield strong incongruence in the phylogeny of eutherian mammals, as revealed by subsampling analyses of loci and taxa, which produced strongly conflicting topologies. In contrast, the coalescent methods, which accommodate gene tree heterogeneity, yield a phylogeny that is robust to variable gene and taxon sampling and is congruent with geographic data. The data also demonstrate that incomplete lineage sorting, a major source of gene tree heterogeneity, is relevant to deep-level phylogenies, such as those among eutherian mammals. Our results firmly place the eutherian root between Atlantogenata and Boreoeutheria and support ungulate polyphyly and a sister-group relationship between Scandentia and Primates. This study demonstrates that the incongruence introduced by concatenation methods is a major cause of long-standing uncertainty in the phylogeny of eutherian mammals, and the same may apply to other clades. Our analyses suggest that such incongruence can be resolved using phylogenomic data and coalescent methods that deal explicitly with gene tree heterogeneity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Concatenation versus coalescence versus "concatalescence".Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):E1179. doi: 10.1073/pnas.1221121110. Epub 2013 Mar 4. Proc Natl Acad Sci U S A. 2013. PMID: 23460702 Free PMC article. No abstract available.
-
Reply to Gatesy and Springer: the multispecies coalescent model can effectively handle recombination and gene tree heterogeneity.Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):E1180. doi: 10.1073/pnas.1300129110. Proc Natl Acad Sci U S A. 2013. PMID: 23650651 Free PMC article. No abstract available.
Similar articles
-
The gene tree delusion.Mol Phylogenet Evol. 2016 Jan;94(Pt A):1-33. doi: 10.1016/j.ympev.2015.07.018. Epub 2015 Jul 31. Mol Phylogenet Evol. 2016. PMID: 26238460
-
Investigating Difficult Nodes in the Placental Mammal Tree with Expanded Taxon Sampling and Thousands of Ultraconserved Elements.Genome Biol Evol. 2017 Sep 1;9(9):2308-2321. doi: 10.1093/gbe/evx168. Genome Biol Evol. 2017. PMID: 28934378 Free PMC article.
-
A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships.Mol Phylogenet Evol. 2003 Aug;28(2):225-40. doi: 10.1016/s1055-7903(03)00118-0. Mol Phylogenet Evol. 2003. PMID: 12878460
-
Implementing and testing the multispecies coalescent model: A valuable paradigm for phylogenomics.Mol Phylogenet Evol. 2016 Jan;94(Pt A):447-62. doi: 10.1016/j.ympev.2015.10.027. Epub 2015 Oct 27. Mol Phylogenet Evol. 2016. PMID: 26518740 Review.
-
Origin of land plants using the multispecies coalescent model.Trends Plant Sci. 2013 Sep;18(9):492-5. doi: 10.1016/j.tplants.2013.04.009. Epub 2013 May 24. Trends Plant Sci. 2013. PMID: 23707196 Review.
Cited by
-
The role of linguistic experience in the development of the consonant bias.Anim Cogn. 2021 May;24(3):419-431. doi: 10.1007/s10071-020-01436-6. Epub 2020 Oct 14. Anim Cogn. 2021. PMID: 33052544
-
Oxytocin and arginine vasopressin receptor evolution: implications for adaptive novelties in placental mammals.Genet Mol Biol. 2016 Oct-Dec;39(4):646-657. doi: 10.1590/1678-4685-GMB-2015-0323. Epub 2016 Aug 8. Genet Mol Biol. 2016. PMID: 27505307 Free PMC article.
-
Evolutionary Models for the Diversification of Placental Mammals Across the KPg Boundary.Front Genet. 2019 Nov 29;10:1241. doi: 10.3389/fgene.2019.01241. eCollection 2019. Front Genet. 2019. PMID: 31850081 Free PMC article.
-
Neural Progenitors in the Developing Neocortex of the Northern Tree Shrew (Tupaia belangeri) Show a Closer Relationship to Gyrencephalic Primates Than to Lissencephalic Rodents.Front Neuroanat. 2018 Apr 19;12:29. doi: 10.3389/fnana.2018.00029. eCollection 2018. Front Neuroanat. 2018. PMID: 29725291 Free PMC article.
-
A multilocus phylogeny reveals deep lineages within African galagids (Primates: Galagidae).BMC Evol Biol. 2014 Apr 2;14:72. doi: 10.1186/1471-2148-14-72. BMC Evol Biol. 2014. PMID: 24694188 Free PMC article.
References
-
- de Queiroz A, Gatesy J. The supermatrix approach to systematics. Trends Ecol Evol. 2007;22(1):34–41. - PubMed
-
- William J, Ballard O. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11:334. - PubMed
-
- Belfiore NM, Liu L, Moritz C. Multilocus phylogenetics of a rapid radiation in the genus Thomomys (Rodentia: Geomyidae) Syst Biol. 2008;57(2):294–310. - PubMed
-
- Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63(1):1–19. - PubMed
-
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol. 2007;56(1):17–24. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

