Comparative mapping in the Fagaceae and beyond with EST-SSRs
- PMID: 22931513
- PMCID: PMC3493355
- DOI: 10.1186/1471-2229-12-153
Comparative mapping in the Fagaceae and beyond with EST-SSRs
Abstract
Background: Genetic markers and linkage mapping are basic prerequisites for comparative genetic analyses, QTL detection and map-based cloning. A large number of mapping populations have been developed for oak, but few gene-based markers are available for constructing integrated genetic linkage maps and comparing gene order and QTL location across related species.
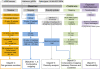
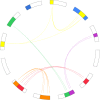
Results: We developed a set of 573 expressed sequence tag-derived simple sequence repeats (EST-SSRs) and located 397 markers (EST-SSRs and genomic SSRs) on the 12 oak chromosomes (2n = 2x = 24) on the basis of Mendelian segregation patterns in 5 full-sib mapping pedigrees of two species: Quercus robur (pedunculate oak) and Quercus petraea (sessile oak). Consensus maps for the two species were constructed and aligned. They showed a high degree of macrosynteny between these two sympatric European oaks. We assessed the transferability of EST-SSRs to other Fagaceae genera and a subset of these markers was mapped in Castanea sativa, the European chestnut. Reasonably high levels of macrosynteny were observed between oak and chestnut. We also obtained diversity statistics for a subset of EST-SSRs, to support further population genetic analyses with gene-based markers. Finally, based on the orthologous relationships between the oak, Arabidopsis, grape, poplar, Medicago, and soybean genomes and the paralogous relationships between the 12 oak chromosomes, we propose an evolutionary scenario of the 12 oak chromosomes from the eudicot ancestral karyotype.
Conclusions: This study provides map locations for a large set of EST-SSRs in two oak species of recognized biological importance in natural ecosystems. This first step toward the construction of a gene-based linkage map will facilitate the assignment of future genome scaffolds to pseudo-chromosomes. This study also provides an indication of the potential utility of new gene-based markers for population genetics and comparative mapping within and beyond the Fagaceae.
Figures







References
-
- Bernardo R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Science. 2008;48(5):1649–1664. doi: 10.2135/cropsci2008.03.0131. - DOI
-
- Troggio M, Malacarne G, Coppola G, Segala C, Dustin A, Cartwright DA, Pindo M, Stefanini M, Mank R, Moroldo M, Morgante M, Grando MS, Velasco R. A Dense Single-Nucleotide Polymorphism-Based Genetic Linkage Map of Grapevine (Vitis vinifera L.) Anchoring Pinot NoirBacterial Artificial Chromosome Contigs. Genetics. 2007;176:2637–2650. doi: 10.1534/genetics.106.067462. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

