Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice
- PMID: 22932389
- PMCID: PMC3523687
- DOI: 10.1038/nature11399
Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice
Abstract
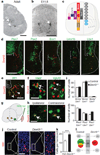
Locomotion in mammals relies on a central pattern-generating circuitry of spinal interneurons established during development that coordinates limb movement. These networks produce left-right alternation of limbs as well as coordinated activation of flexor and extensor muscles. Here we show that a premature stop codon in the DMRT3 gene has a major effect on the pattern of locomotion in horses. The mutation is permissive for the ability to perform alternate gaits and has a favourable effect on harness racing performance. Examination of wild-type and Dmrt3-null mice demonstrates that Dmrt3 is expressed in the dI6 subdivision of spinal cord neurons, takes part in neuronal specification within this subdivision, and is critical for the normal development of a coordinated locomotor network controlling limb movements. Our discovery positions Dmrt3 in a pivotal role for configuring the spinal circuits controlling stride in vertebrates. The DMRT3 mutation has had a major effect on the diversification of the domestic horse, as the altered gait characteristics of a number of breeds apparently require this mutation.
Figures


References
-
- Grillner S. Biological pattern generation: the cellular and computational logic of networks in motion. Neuron. 2006;52:751–766. - PubMed
-
- Kullander K. Genetics moving to neuronal networks. Trends Neurosci. 2005;28:239–247. - PubMed
-
- Albertsdóttir E, Eriksson S, Sigurdsson A, Arnason T. Genetic analysis of ‘breeding field test status’ in Icelandic horses. J. Anim. Breed. Genet. 2011;128:124–132. - PubMed
-
- Hong CS, Park BY, Saint-Jeannet JP. The function of Dmrt genes in vertebrate development: it is not just about sex. Dev. Biol. 2007;310:1–9. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

