Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis
- PMID: 22932667
- PMCID: PMC5081094
- DOI: 10.1158/1078-0432.CCR-12-1807
Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis
Abstract
Purpose: There is a need for improved subclassification of urothelial carcinoma (UC) at diagnosis. A major aim of this study was to search for novel genomic subgroups.
Experimental design: We assessed 160 tumors for genome-wide copy number alterations and mutation in genes implicated in UC. These comprised all tumor grades and stages and included 49 high-grade stage T1 (T1G3) tumors.
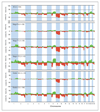
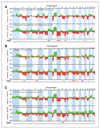
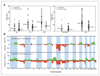
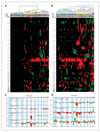
Results: Our findings point to the existence of genomic subclasses of the "gold-standard" grade/stage groups. The T1G3 tumors separated into 3 major subgroups that differed with respect to the type and number of copy number events and to FGFR3 and TP53 mutation status. We also identified novel regions of copy number alteration, uncovered relationships between molecular events, and elucidated relationships between molecular events and clinico-pathologic features. FGFR3 mutant tumors were more chromosomally stable than their wild-type counterparts and a mutually exclusive relationship between FGFR3 mutation and overrepresentation of 8q was observed in non-muscle-invasive tumors. In muscle-invasive (MI) tumors, metastasis was positively associated with losses of regions on 10q (including PTEN), 16q and 22q, and gains on 10p, 11q, 12p, 19p, and 19q. Concomitant copy number alterations positively associated with TP53 mutation in MI tumors were losses on 16p, 2q, 4q, 11p, 10q, 13q, 14q, 16q, and 19p, and gains on 1p, 8q, 10q, and 12q. Significant complexity was revealed in events affecting chromosome 9.
Conclusions: These findings may lead to improved biologic understanding and the development of prognostic biomarkers. Novel regions of copy number alteration may reveal potential therapeutic targets.
©2012 AACR.
Conflict of interest statement
of Potential Confiicts of Interest No potential conflicts of interest were disclosed.
Figures
References
-
- Montie JE, Clark PE, Eisenberger MA, El-Galley R, Greenberg RE, Herr HW, et al. Bladder cancer. J Natl Compr Canc Netw. 2009;7:8–39. - PubMed
-
- López-Knowles E, Hernández S, Malats N, Kogevinas M, Lloreta J, Carrato A, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66:7401–4. - PubMed
-
- Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5:713–25. - PubMed
-
- Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27:361–73. - PubMed
-
- Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–81. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

