Epitope mapping of broadly neutralizing HIV-2 human monoclonal antibodies
- PMID: 22933274
- PMCID: PMC3486499
- DOI: 10.1128/JVI.01632-12
Epitope mapping of broadly neutralizing HIV-2 human monoclonal antibodies
Abstract
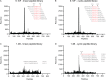
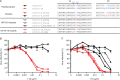
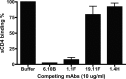
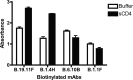
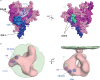
Recent studies have shown that natural infection by HIV-2 leads to the elicitation of high titers of broadly neutralizing antibodies (NAbs) against primary HIV-2 strains (T. I. de Silva, et al., J. Virol. 86:930-946, 2012; R. Kong, et al., J. Virol. 86:947-960, 2012; G. Ozkaya Sahin, et al., J. Virol. 86:961-971, 2012). Here, we describe the envelope (Env) binding and neutralization properties of 15 anti-HIV-2 human monoclonal antibodies (MAbs), 14 of which were newly generated from 9 chronically infected subjects. All 15 MAbs bound specifically to HIV-2 gp120 monomers and neutralized heterologous primary virus strains HIV-2(7312A) and HIV-2(ST). Ten of 15 MAbs neutralized a third heterologous primary virus strain, HIV-2(UC1). The median 50% inhibitory concentrations (IC(50)s) for these MAbs were surprisingly low, ranging from 0.007 to 0.028 μg/ml. Competitive Env binding studies revealed three MAb competition groups: CG-I, CG-II, and CG-III. Using peptide scanning, site-directed mutagenesis, chimeric Env constructions, and single-cycle virus neutralization assays, we mapped the epitope of CG-I antibodies to a linear region in variable loop 3 (V3), the epitope of CG-II antibodies to a conformational region centered on the carboxy terminus of V4, and the epitope(s) of CG-III antibodies to conformational regions associated with CD4- and coreceptor-binding sites. HIV-2 Env is thus highly immunogenic in vivo and elicits antibodies having diverse epitope specificities, high potency, and wide breadth. In contrast to the HIV-1 Env trimer, which is generally well shielded from antibody binding and neutralization, HIV-2 is surprisingly vulnerable to broadly reactive NAbs. The availability of 15 human MAbs targeting diverse HIV-2 Env epitopes can facilitate comparative studies of HIV/SIV Env structure, function, antigenicity, and immunogenicity.
Figures

References
-
- Binley JM, et al. 1996. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res. Hum. Retroviruses 12:911–924 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI094604/AI/NIAID NIH HHS/United States
- R33 AI087383/AI/NIAID NIH HHS/United States
- U01 AI067854/AI/NIAID NIH HHS/United States
- R01 AI087181/AI/NIAID NIH HHS/United States
- G0801751/MRC_/Medical Research Council/United Kingdom
- AI88564/AI/NIAID NIH HHS/United States
- AI87383/AI/NIAID NIH HHS/United States
- U19 AI067854/AI/NIAID NIH HHS/United States
- MC_UP_A900_1121/MRC_/Medical Research Council/United Kingdom
- ImNIH/Intramural NIH HHS/United States
- R21 AI087383/AI/NIAID NIH HHS/United States
- R01 AI084830/AI/NIAID NIH HHS/United States
- MC_U137884180/MRC_/Medical Research Council/United Kingdom
- P01 AI088564/AI/NIAID NIH HHS/United States
- AI67854/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

