The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae
- PMID: 22933288
- PMCID: PMC3486445
- DOI: 10.1128/JVI.01873-12
The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae
Abstract
The family Baculoviridae is a large group of insect viruses containing circular double-stranded DNA genomes of 80 to 180 kbp, which have broad biotechnological applications. A key feature to understand and manipulate them is the recognition of orthology. However, the differences in gene contents and evolutionary distances among the known members of this family make it difficult to assign sequence orthology. In this study, the genome sequences of 58 baculoviruses were analyzed, with the aim to detect previously undescribed core genes because of their remote homology. A routine based on Multi PSI-Blast/tBlastN and Multi HaMStR allowed us to detect 31 of 33 accepted core genes and 4 orthologous sequences in the Baculoviridae which were not described previously. Our results show that the ac53, ac78, ac101 (p40), and ac103 (p48) genes have orthologs in all genomes and should be considered core genes. Accordingly, there are 37 orthologous genes in the family Baculoviridae.
Figures
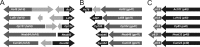



References
-
- Acharya A, Gopinathan K. 2002. Transcriptional analysis and preliminary characterization of ORF Bm42 from Bombyx mori nucleopolyhedrovirus. Virology 299:213–224 - PubMed
-
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 - PubMed
-
- Blissard GW, Rohrmann GF. 1990. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 35:127–155 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

