Single-neuron RNA-Seq: technical feasibility and reproducibility
- PMID: 22934102
- PMCID: PMC3407998
- DOI: 10.3389/fgene.2012.00124
Single-neuron RNA-Seq: technical feasibility and reproducibility
Erratum in
- Front Genet. 2013;4:23
- Front Genet. 2013;4:69
Abstract
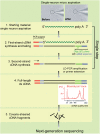
Understanding brain function involves improved knowledge about how the genome specifies such a large diversity of neuronal types. Transcriptome analysis of single neurons has been previously described using gene expression microarrays. Using high-throughput transcriptome sequencing (RNA-Seq), we have developed a method to perform single-neuron RNA-Seq. Following electrophysiology recording from an individual neuron, total RNA was extracted by aspirating the cellular contents into a fine glass electrode tip. The mRNAs were reverse transcribed and amplified to construct a single-neuron cDNA library, and subsequently subjected to high-throughput sequencing. This approach was applied to both individual neurons cultured from embryonic mouse hippocampus, as well as neocortical neurons from live brain slices. We found that the average pairwise Spearman's rank correlation coefficient of gene expression level expressed as RPKM (reads per kilobase of transcript per million mapped reads) was 0.51 between five cultured neuronal cells, whereas the same measure between three cortical layer 5 neurons in situ was 0.25. The data suggest that there may be greater heterogeneity of the cortical neurons, as compared to neurons in vitro. The results demonstrate the technical feasibility and reproducibility of RNA-Seq in capturing a part of the transcriptome landscape of single neurons, and confirmed that morphologically identical neurons, even from the same region, have distinct gene expression patterns.
Keywords: RNA-Seq; cell culture; electrophysiology; gene expression; neuron; transcriptome.
Figures


Similar articles
-
Microfluidic single-cell whole-transcriptome sequencing.Proc Natl Acad Sci U S A. 2014 May 13;111(19):7048-53. doi: 10.1073/pnas.1402030111. Epub 2014 Apr 29. Proc Natl Acad Sci U S A. 2014. PMID: 24782542 Free PMC article.
-
Single read and paired end mRNA-Seq Illumina libraries from 10 nanograms total RNA.J Vis Exp. 2011 Oct 27;(56):e3340. doi: 10.3791/3340. J Vis Exp. 2011. PMID: 22064688 Free PMC article.
-
Differing molecular response of young and advanced maternal age human oocytes to IVM.Hum Reprod. 2017 Nov 1;32(11):2199-2208. doi: 10.1093/humrep/dex284. Hum Reprod. 2017. PMID: 29025019 Free PMC article.
-
Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols.RNA. 2020 Aug;26(8):903-909. doi: 10.1261/rna.074922.120. Epub 2020 Apr 13. RNA. 2020. PMID: 32284352 Free PMC article. Review.
-
Introduction to sequencing the brain transcriptome.Int Rev Neurobiol. 2014;116:1-19. doi: 10.1016/B978-0-12-801105-8.00001-1. Int Rev Neurobiol. 2014. PMID: 25172469 Free PMC article. Review.
Cited by
-
Single neurons needed for brain asymmetry studies.Front Genet. 2014 Jan 16;4:311. doi: 10.3389/fgene.2013.00311. eCollection 2013. Front Genet. 2014. PMID: 24474957 Free PMC article. No abstract available.
-
Kcnn2 blockade reverses learning deficits in a mouse model of fetal alcohol spectrum disorders.Nat Neurosci. 2020 Apr;23(4):533-543. doi: 10.1038/s41593-020-0592-z. Epub 2020 Mar 16. Nat Neurosci. 2020. PMID: 32203497 Free PMC article.
-
SINCERA: A Pipeline for Single-Cell RNA-Seq Profiling Analysis.PLoS Comput Biol. 2015 Nov 24;11(11):e1004575. doi: 10.1371/journal.pcbi.1004575. eCollection 2015 Nov. PLoS Comput Biol. 2015. PMID: 26600239 Free PMC article.
-
Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes.Nat Biotechnol. 2016 Feb;34(2):175-183. doi: 10.1038/nbt.3443. Epub 2015 Dec 21. Nat Biotechnol. 2016. PMID: 26689544 Free PMC article.
-
A RNA-Seq Analysis of the Rat Supraoptic Nucleus Transcriptome: Effects of Salt Loading on Gene Expression.PLoS One. 2015 Apr 21;10(4):e0124523. doi: 10.1371/journal.pone.0124523. eCollection 2015. PLoS One. 2015. PMID: 25897513 Free PMC article.
References
-
- Birney E., Andrews T. D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Down T., Eyras E., Fernandez-Suarez X. M., Gane P., Gibbins B., Gilbert J., Hammond M., Hotz H. R., Iyer V., Jekosch K., Kahari A., Kasprzyk A., Keefe D., Keenan S., Lehvaslaiho H., Mcvicker G., Melsopp C., Meidl P., Mongin E., Pettett R., Potter S., Proctor G., Rae M., Searle S., Slater G., Smedley D., Smith J., Spooner W., Stabenau A., Stalker J., Storey R., Ureta-Vidal A., Woodwark K. C., Cameron G., Durbin R., Cox A., Hubbard T., Clamp M. (2004). An overview of Ensembl. Genome Res. 14 925–928 - PMC - PubMed
-
- Bota M., Dong H. W., Swanson L. W. (2003). From gene networks to brain networks. Nat. Neurosci. 6 795–799 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

