Development of Therapeutic-Grade Small Interfering RNAs by Chemical Engineering
- PMID: 22934103
- PMCID: PMC3422727
- DOI: 10.3389/fgene.2012.00154
Development of Therapeutic-Grade Small Interfering RNAs by Chemical Engineering
Abstract
Recent successes in clinical trials have provided important proof of concept that small interfering RNAs (siRNAs) indeed constitute a new promising class of therapeutics. Although great efforts are still needed to ensure efficient means of delivery in vivo, the siRNA molecule itself has been successfully engineered by chemical modification to meet initial challenges regarding specificity, stability, and immunogenicity. To date, a great wealth of siRNA architectures and types of chemical modification are available for promoting safe siRNA-mediated gene silencing in vivo and, consequently, the choice of design and modification types can be challenging to individual experimenters. Here we review the literature and devise how to improve siRNA performance by structural design and specific chemical modification to ensure potent and specific gene silencing without unwarranted side-effects and hereby complement the ongoing efforts to improve cell targeting and delivery by other carrier molecules.
Keywords: LNA; OMe; RNAi; chemical modification; immunogenicity; off-target effect; siRNA; siRNA therapeutic.
Figures
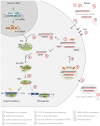
 and, although degradation rates are much lower in the cytoplasm, siRNAs stabilization by modification is suggested to also enhance intracellular availability and silencing persistence
and, although degradation rates are much lower in the cytoplasm, siRNAs stabilization by modification is suggested to also enhance intracellular availability and silencing persistence  . Also, extracellular siRNAs can be rapidly cleared from the body, e.g., by renal filtration
. Also, extracellular siRNAs can be rapidly cleared from the body, e.g., by renal filtration  and can induce innate immune responses via TLR3 in certain cells
and can induce innate immune responses via TLR3 in certain cells  . Delivery across the target cell membrane
. Delivery across the target cell membrane  and endosomal release of endocytosed
and endosomal release of endocytosed  are currently the main bottlenecks in siRNA applications in vivo and siRNAs may induce TLR7/8-mediated immune-responses upon endosome retention in immune cells
are currently the main bottlenecks in siRNA applications in vivo and siRNAs may induce TLR7/8-mediated immune-responses upon endosome retention in immune cells  . Also, all cells can respond to foreign cytoplasmic RNA via the PRRs, PKR, RIG-I, and Mda5
. Also, all cells can respond to foreign cytoplasmic RNA via the PRRs, PKR, RIG-I, and Mda5  . siRNA may disturb natural miRNA pathways, that processes nuclear pri-miRNA transcripts (dark gray circle) via a pre-miRNA intermediate and miRNA duplex into a single-stranded miRNA in RISC, by direct competition for RISC loading
. siRNA may disturb natural miRNA pathways, that processes nuclear pri-miRNA transcripts (dark gray circle) via a pre-miRNA intermediate and miRNA duplex into a single-stranded miRNA in RISC, by direct competition for RISC loading  or by clotting the pathway due to slow siRNA processing and turnover
or by clotting the pathway due to slow siRNA processing and turnover  . Finally, all siRNA will trigger miRNA-like off-targeting effects on unintended targets upon base-pairing of the guide strand seed region and positions within mRNA 3′ UTRs leading to transcript destabilization and/or translational repression
. Finally, all siRNA will trigger miRNA-like off-targeting effects on unintended targets upon base-pairing of the guide strand seed region and positions within mRNA 3′ UTRs leading to transcript destabilization and/or translational repression  . Please refer to main text for more detail.
. Please refer to main text for more detail.


Similar articles
-
Engineering small interfering RNAs by strategic chemical modification.Methods Mol Biol. 2013;942:87-109. doi: 10.1007/978-1-62703-119-6_5. Methods Mol Biol. 2013. PMID: 23027047
-
Chemical and structural modifications of RNAi therapeutics.Adv Drug Deliv Rev. 2016 Sep 1;104:16-28. doi: 10.1016/j.addr.2015.10.015. Epub 2015 Nov 5. Adv Drug Deliv Rev. 2016. PMID: 26549145 Review.
-
Development of RNA interference-based therapeutics and application of multi-target small interfering RNAs.Nucleic Acid Ther. 2014 Aug;24(4):302-12. doi: 10.1089/nat.2014.0480. Epub 2014 May 5. Nucleic Acid Ther. 2014. PMID: 24796432 Review.
-
Re-Engineering RNA Molecules into Therapeutic Agents.Acc Chem Res. 2019 Apr 16;52(4):1036-1047. doi: 10.1021/acs.accounts.8b00650. Epub 2019 Mar 26. Acc Chem Res. 2019. PMID: 30912917
-
Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo.Mol Cancer Ther. 2007 Mar;6(3):833-43. doi: 10.1158/1535-7163.MCT-06-0195. Mol Cancer Ther. 2007. PMID: 17363479
Cited by
-
SMEpred workbench: A web server for predicting efficacy of chemicallymodified siRNAs.RNA Biol. 2016 Nov;13(11):1144-1151. doi: 10.1080/15476286.2016.1229733. Epub 2016 Sep 7. RNA Biol. 2016. PMID: 27603513 Free PMC article.
-
Synthesis and evaluation of modified siRNA molecules containing a novel glucose derivative.RSC Adv. 2021 Mar 1;11(16):9285-9289. doi: 10.1039/d1ra00922b. eCollection 2021 Mar 1. RSC Adv. 2021. PMID: 35423452 Free PMC article.
-
Multifunctional RNA nanoparticles.Nano Lett. 2014 Oct 8;14(10):5662-71. doi: 10.1021/nl502385k. Epub 2014 Sep 30. Nano Lett. 2014. PMID: 25267559 Free PMC article.
-
A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution.Nanomedicine. 2020 Jan;23:102094. doi: 10.1016/j.nano.2019.102094. Epub 2019 Oct 25. Nanomedicine. 2020. PMID: 31669854 Free PMC article.
-
Expression and Purification of tRNA/ pre-miRNA-Based Recombinant Noncoding RNAs.Methods Mol Biol. 2021;2323:249-265. doi: 10.1007/978-1-0716-1499-0_18. Methods Mol Biol. 2021. PMID: 34086286 Free PMC article.
References
-
- Abe N., Abe H., Nagai C., Harada M., Hatakeyama H., Harashima H., Ohshiro T., Nishihara M., Furukawa K., Maeda M., Tsuneda S., Ito Y. (2011). Synthesis, structure, and biological activity of dumbbell-shaped nanocircular RNAs for RNA interference. Bioconjug. Chem. 22, 2082–209210.1021/bc2003154 - DOI - PubMed
-
- Allerson C. R., Sioufi N., Jarres R., Prakash T. P., Naik N., Berdeja A., Wanders L., Griffey R. H., Swayze E. E., Bhat B. (2005). Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 48, 901–90410.1021/jm049167j - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

