Metabolic context regulates distinct hypothalamic transcriptional responses to antiaging interventions
- PMID: 22934110
- PMCID: PMC3427989
- DOI: 10.1155/2012/732975
Metabolic context regulates distinct hypothalamic transcriptional responses to antiaging interventions
Abstract
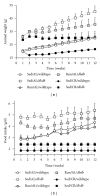
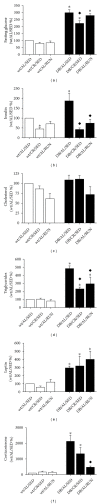
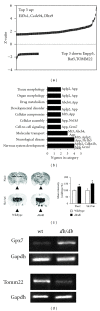
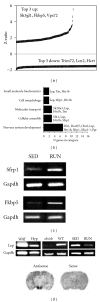
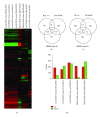
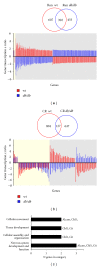
The hypothalamus is an essential relay in the neural circuitry underlying energy metabolism that needs to continually adapt to changes in the energetic environment. The neuroendocrine control of food intake and energy expenditure is associated with, and likely dependent upon, hypothalamic plasticity. Severe disturbances in energy metabolism, such as those that occur in obesity, are therefore likely to be associated with disruption of hypothalamic transcriptomic plasticity. In this paper, we investigated the effects of two well-characterized antiaging interventions, caloric restriction and voluntary wheel running, in two distinct physiological paradigms, that is, diabetic (db/db) and nondiabetic wild-type (C57/Bl/6) animals to investigate the contextual sensitivity of hypothalamic transcriptomic responses. We found that, both quantitatively and qualitatively, caloric restriction and physical exercise were associated with distinct transcriptional signatures that differed significantly between diabetic and non-diabetic mice. This suggests that challenges to metabolic homeostasis regulate distinct hypothalamic gene sets in diabetic and non-diabetic animals. A greater understanding of how genetic background contributes to hypothalamic response mechanisms could pave the way for the development of more nuanced therapeutics for the treatment of metabolic disorders that occur in diverse physiological backgrounds.
Figures



Similar articles
-
Hypothalamic C2-domain protein involved in MC4R trafficking and control of energy balance.Metabolism. 2020 Jan;102:153990. doi: 10.1016/j.metabol.2019.153990. Epub 2019 Oct 27. Metabolism. 2020. PMID: 31666192
-
Hypothalamic Dysfunction in Obesity and Metabolic Disorders.Adv Neurobiol. 2017;19:73-116. doi: 10.1007/978-3-319-63260-5_4. Adv Neurobiol. 2017. PMID: 28933062 Review.
-
A spontaneous leptin receptor point mutation causes obesity and differentially affects leptin signaling in hypothalamic nuclei resulting in metabolic dysfunctions distinct from db/db mice.Mol Metab. 2019 Jul;25:131-141. doi: 10.1016/j.molmet.2019.04.010. Epub 2019 Apr 25. Mol Metab. 2019. PMID: 31076350 Free PMC article.
-
Sex-Biased Physiological Roles of NPFF1R, the Canonical Receptor of RFRP-3, in Food Intake and Metabolic Homeostasis Revealed by its Congenital Ablation in mice.Metabolism. 2018 Oct;87:87-97. doi: 10.1016/j.metabol.2018.07.003. Epub 2018 Jul 31. Metabolism. 2018. PMID: 30075164
-
Effects of physical exercise on food intake and body weight: Role of dorsomedial hypothalamic signaling.Physiol Behav. 2018 Aug 1;192:59-63. doi: 10.1016/j.physbeh.2018.03.018. Epub 2018 Mar 17. Physiol Behav. 2018. PMID: 29559223 Review.
Cited by
-
Gene co-expression analyses of health(span) across multiple species.NAR Genom Bioinform. 2022 Nov 29;4(4):lqac083. doi: 10.1093/nargab/lqac083. eCollection 2022 Dec. NAR Genom Bioinform. 2022. PMID: 36458022 Free PMC article.
-
Models and mechanisms for hippocampal dysfunction in obesity and diabetes.Neuroscience. 2015 Nov 19;309:125-39. doi: 10.1016/j.neuroscience.2015.04.045. Epub 2015 Apr 28. Neuroscience. 2015. PMID: 25934036 Free PMC article. Review.
-
The Relaxin-3 Receptor, RXFP3, Is a Modulator of Aging-Related Disease.Int J Mol Sci. 2022 Apr 15;23(8):4387. doi: 10.3390/ijms23084387. Int J Mol Sci. 2022. PMID: 35457203 Free PMC article. Review.
-
Hippocampal brain-derived neurotrophic factor determines recruitment of anatomically connected networks after stress in diabetic mice.Hippocampus. 2018 Dec;28(12):900-912. doi: 10.1002/hipo.23018. Epub 2018 Nov 6. Hippocampus. 2018. PMID: 30098276 Free PMC article.
-
Longitudinal analysis of calorie restriction on rat taste bud morphology and expression of sweet taste modulators.J Gerontol A Biol Sci Med Sci. 2014 May;69(5):532-44. doi: 10.1093/gerona/glt129. Epub 2013 Sep 28. J Gerontol A Biol Sci Med Sci. 2014. PMID: 24077597 Free PMC article.
References
-
- Jackson K, Vieira Silva HM, Zhang W, Michelini LC, Stern JE. Exercise training differentially affects intrinsic excitability of autonomic and neuroendocrine neurons in the hypothalamic paraventricular nucleus. Journal of Neurophysiology. 2005;94(5):3211–3220. - PubMed
-
- Sternson SM, Shepherd GMG, Friedman JM. Topographic mapping of VMH arcuate nucleus microcircuits and their reorganization by fasting. Nature Neuroscience. 2005;8(10):1356–1363. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

