Host cell cytotoxicity and cytoskeleton disruption by CerADPr, an ADP-ribosyltransferase of Bacillus cereus G9241
- PMID: 22934824
- PMCID: PMC3661007
- DOI: 10.1021/bi300692g
Host cell cytotoxicity and cytoskeleton disruption by CerADPr, an ADP-ribosyltransferase of Bacillus cereus G9241
Abstract
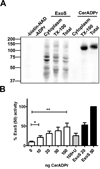
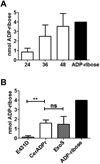
Bacillus cereus G9241 was isolated from a welder suffering from an anthrax-like inhalation illness. B. cereus G9241 encodes two megaplasmids, pBCXO1 and pBC210, which are analogous to the toxin- and capsule-encoding virulence plasmids of Bacillus anthracis. Protein modeling predicted that the pBC210 LF homologue contained an ADP-ribosyltransferase (ADPr) domain. This putative bacterial ADP-ribosyltransferase domain was denoted CerADPr. Iterative modeling showed that CerADPr possessed several conserved ADP-ribosyltransferase features, including an α-3 helix, an ADP-ribosyltransferase turn-turn loop, and a "Gln-XXX-Glu" motif. CerADPr ADP-ribosylated an ~120 kDa protein in HeLa cell lysates and intact cells. EGFP-CerADPr rounded HeLa cells, elicited cytoskeletal changes, and yielded a cytotoxic phenotype, indicating that CerADPr disrupts cytoskeletal signaling. CerADPr(E431D) did not possess ADP-ribosyltransferase or NAD glycohydrolase activities and did not elicit a phenotype in HeLa cells, implicating Glu431 as a catalytic residue. These experiments identify CerADPr as a cytotoxic ADP-ribosyltransferase that disrupts the host cytoskeleton.
Figures



Similar articles
-
Certhrax Is an Antivirulence Factor for the Anthrax-Like Organism Bacillus cereus Strain G9241.Infect Immun. 2018 May 22;86(6):e00207-18. doi: 10.1128/IAI.00207-18. Print 2018 Jun. Infect Immun. 2018. PMID: 29610258 Free PMC article.
-
Bacillus cereus G9241 makes anthrax toxin and capsule like highly virulent B. anthracis Ames but behaves like attenuated toxigenic nonencapsulated B. anthracis Sterne in rabbits and mice.Infect Immun. 2011 Aug;79(8):3012-9. doi: 10.1128/IAI.00205-11. Epub 2011 May 16. Infect Immun. 2011. PMID: 21576337 Free PMC article.
-
Bacillus cereus Certhrax ADP-ribosylates vinculin to disrupt focal adhesion complexes and cell adhesion.J Biol Chem. 2014 Apr 11;289(15):10650-10659. doi: 10.1074/jbc.M113.500710. Epub 2014 Feb 26. J Biol Chem. 2014. PMID: 24573681 Free PMC article.
-
Studies on the active-site structure of C3-like exoenzymes: involvement of glutamic acid in catalysis of ADP-ribosylation.Biochimie. 1995;77(5):326-32. doi: 10.1016/0300-9084(96)88142-9. Biochimie. 1995. PMID: 8527485 Review.
-
Pseudomonas aeruginosa ExoS and ExoT.Rev Physiol Biochem Pharmacol. 2004;152:79-92. doi: 10.1007/s10254-004-0031-7. Epub 2004 Aug 24. Rev Physiol Biochem Pharmacol. 2004. PMID: 15375697 Review.
Cited by
-
Expression and contribution to virulence of each polysaccharide capsule of Bacillus cereus strain G9241.PLoS One. 2018 Aug 22;13(8):e0202701. doi: 10.1371/journal.pone.0202701. eCollection 2018. PLoS One. 2018. PMID: 30133532 Free PMC article.
-
Vaccine protection against Bacillus cereus-mediated respiratory anthrax-like disease in mice.Infect Immun. 2013 Mar;81(3):1008-17. doi: 10.1128/IAI.01346-12. Epub 2013 Jan 14. Infect Immun. 2013. PMID: 23319564 Free PMC article.
-
You Can't B. cereus - A Review of Bacillus cereus Strains That Cause Anthrax-Like Disease.Front Microbiol. 2020 Aug 19;11:1731. doi: 10.3389/fmicb.2020.01731. eCollection 2020. Front Microbiol. 2020. PMID: 32973690 Free PMC article. Review.
-
The Bacillus cereus Group: Bacillus Species with Pathogenic Potential.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0032-2018. doi: 10.1128/microbiolspec.GPP3-0032-2018. Microbiol Spectr. 2019. PMID: 31111815 Free PMC article. Review.
-
The roles of AtxA orthologs in virulence of anthrax-like Bacillus cereus G9241.Mol Microbiol. 2016 Nov;102(4):545-561. doi: 10.1111/mmi.13478. Epub 2016 Sep 4. Mol Microbiol. 2016. PMID: 27490458 Free PMC article.
References
-
- Moss J, et al. NAD-dependent ADP-ribosylation of arginine and proteins by Escherichia coli heat-labile enterotoxin. Journal of Biological Chemistry. 1979;254(14):6270–6272. - PubMed
-
- Collier RJ. Effect of diphtheria toxin on protein synthesis: Inactivation of one of the transfer factors. Journal of Molecular Biology. 1967;25(1):83–98. - PubMed
-
- Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. Journal of Biological Chemistry. 1980;255(22):10717–10720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

