Mass spectrometry (LC-MS/MS) identified proteomic biosignatures of breast cancer in proximal fluid
- PMID: 22934887
- PMCID: PMC3521600
- DOI: 10.1021/pr300606e
Mass spectrometry (LC-MS/MS) identified proteomic biosignatures of breast cancer in proximal fluid
Abstract
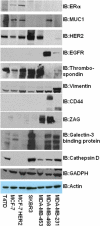
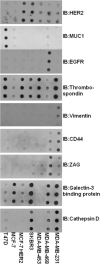
We have begun an early phase of biomarker discovery in three clinically important types of breast cancer using a panel of human cell lines: HER2 positive, hormone receptor positive and HER2 negative, and triple negative (HER2-, ER-, PR-). We identified and characterized the most abundant secreted, sloughed, or leaked proteins released into serum free media from these breast cancer cell lines using a combination of protein fractionation methods before LC-MS/MS mass spectrometry analysis. A total of 249 proteins were detected in the proximal fluid of 7 breast cancer cell lines. The expression of a selected group of high abundance and/or breast cancer-specific potential biomarkers including thromobospondin 1, galectin-3 binding protein, cathepsin D, vimentin, zinc-α2-glycoprotein, CD44, and EGFR from the breast cancer cell lines and in their culture media were further validated by Western blot analysis. Interestingly, mass spectrometry identified a cathepsin D protein single-nucleotide polymorphism (SNP) by alanine to valine replacement from the MCF-7 breast cancer cell line. Comparison of each cell line media proteome displayed unique and consistent biosignatures regardless of the individual group classifications, demonstrating the potential for stratification of breast cancer. On the basis of the cell line media proteome, predictive Tree software was able to categorize each cell line as HER2 positive, HER2 negative, and hormone receptor positive and triple negative based on only two proteins, muscle fructose 1,6-bisphosphate aldolase and keratin 19. In addition, the predictive Tree software clearly identified MCF-7 cell line overexpresing the HER2 receptor with the SNP cathepsin D biomarker.
Figures




Similar articles
-
Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer.J Proteome Res. 2007 Aug;6(8):2993-3002. doi: 10.1021/pr060629m. Epub 2007 Jul 4. J Proteome Res. 2007. PMID: 17608509 Free PMC article.
-
Quantitative proteomic analysis of HER2 normal and overexpressing MCF-7 breast cancer cells revealed proteomic changes accompanied with HER2 gene amplification.J Proteomics. 2013 Oct 8;91:200-9. doi: 10.1016/j.jprot.2013.06.034. Epub 2013 Jul 11. J Proteomics. 2013. PMID: 23851309
-
Comparative membrane proteomics analyses of breast cancer cell lines to understand the molecular mechanism of breast cancer brain metastasis.Electrophoresis. 2017 Sep;38(17):2124-2134. doi: 10.1002/elps.201700027. Epub 2017 Jul 5. Electrophoresis. 2017. PMID: 28523741
-
The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context.Biochim Biophys Acta. 2013 Nov;1834(11):2242-58. doi: 10.1016/j.bbapap.2013.01.029. Epub 2013 Jan 31. Biochim Biophys Acta. 2013. PMID: 23376433 Review.
-
The clinical impact of recent advances in LC-MS for cancer biomarker discovery and verification.Expert Rev Proteomics. 2016;13(1):99-114. doi: 10.1586/14789450.2016.1122529. Epub 2015 Dec 19. Expert Rev Proteomics. 2016. PMID: 26581546 Free PMC article. Review.
Cited by
-
Quantitative proteomic analysis of pancreatic cyst fluid proteins associated with malignancy in intraductal papillary mucinous neoplasms.Clin Proteomics. 2018 Apr 18;15:17. doi: 10.1186/s12014-018-9193-1. eCollection 2018. Clin Proteomics. 2018. PMID: 29713252 Free PMC article.
-
Proteomics-Based Identification of Dysregulated Proteins and Biomarker Discovery in Invasive Ductal Carcinoma, the Most Common Breast Cancer Subtype.Proteomes. 2023 Apr 3;11(2):13. doi: 10.3390/proteomes11020013. Proteomes. 2023. PMID: 37092454 Free PMC article. Review.
-
Integration of Breast Cancer Secretomes with Clinical Data Elucidates Potential Serum Markers for Disease Detection, Diagnosis, and Prognosis.PLoS One. 2016 Jun 29;11(6):e0158296. doi: 10.1371/journal.pone.0158296. eCollection 2016. PLoS One. 2016. PMID: 27355404 Free PMC article.
-
Proteomics-Based Identification of Dysregulated Proteins in Breast Cancer.Proteomes. 2022 Oct 21;10(4):35. doi: 10.3390/proteomes10040035. Proteomes. 2022. PMID: 36278695 Free PMC article. Review.
-
Plasma Peptidome Pattern of Breast Cancer Using Magnetic Beads-Based Plasma Fractionation and MALDI-TOF MS: A Case Control Study in Egypt.Asian Pac J Cancer Prev. 2019 Jan 25;20(1):175-184. doi: 10.31557/APJCP.2019.20.1.175. Asian Pac J Cancer Prev. 2019. PMID: 30678429 Free PMC article.
References
-
- Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nature reviews. 2003;3(4):243–52. - PubMed
-
- Armstrong K, Moye E, Williams S, Berlin JA, Reynolds EE. Screening mammography in women 40 to 49 years of age: a systematic review for the American College of Physicians. Annals of internal medicine. 2007;146(7):516–26. - PubMed
-
- Glick SJ, Breast CT. Annu Rev Biomed Eng. 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

