The prolyl-isomerase Pin1 activates the mitochondrial death program of p53
- PMID: 22935610
- PMCID: PMC3554345
- DOI: 10.1038/cdd.2012.112
The prolyl-isomerase Pin1 activates the mitochondrial death program of p53
Abstract
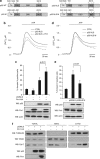
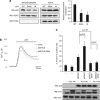
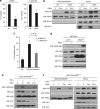
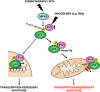
In response to intense stress, the tumor protein p53 (p53) tumor suppressor rapidly mounts a direct mitochondrial death program that precedes transcription-mediated apoptosis. By eliminating severely damaged cells, this pathway contributes to tumor suppression as well as to cancer cell killing induced by both genotoxic drugs and non-genotoxic p53-reactivating molecules. Here we have explored the role had in this pathway by the prolyl-isomerase Pin1 (peptidylprolyl cis/trans isomerase, NIMA-interacting 1), a crucial transducer of p53's phosphorylation into conformational changes unleashing its pro-apoptotic activity. We show that Pin1 promotes stress-induced localization of p53 to mitochondria both in vitro and in vivo. In particular, we demonstrate that upon stress-induced phosphorylation of p53 on Ser46 by homeodomain interacting protein kinase 2, Pin1 stimulates its mitochondrial trafficking signal, that is, monoubiquitination. This pathway is induced also by the p53-activating molecule RITA, and we demonstrate the strong requirement of Pin1 for the induction of mitochondrial apoptosis by this compound. These findings have significant implications for treatment of p53-expressing tumors and for prospective use of p53-activating compounds in clinics.
Figures


Similar articles
-
The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP.Nat Struct Mol Biol. 2007 Oct;14(10):912-20. doi: 10.1038/nsmb1306. Epub 2007 Sep 30. Nat Struct Mol Biol. 2007. PMID: 17906639
-
Ser46 phosphorylation of p53 is an essential event in prolyl-isomerase Pin1-mediated p53-independent apoptosis in response to heat stress.Cell Death Dis. 2019 Feb 4;10(2):96. doi: 10.1038/s41419-019-1316-8. Cell Death Dis. 2019. PMID: 30718466 Free PMC article.
-
Ser46 phosphorylation and prolyl-isomerase Pin1-mediated isomerization of p53 are key events in p53-dependent apoptosis induced by mutant huntingtin.Proc Natl Acad Sci U S A. 2011 Nov 1;108(44):17979-84. doi: 10.1073/pnas.1106198108. Epub 2011 Oct 19. Proc Natl Acad Sci U S A. 2011. PMID: 22011578 Free PMC article.
-
Interaction of p53 with prolyl isomerases: Healthy and unhealthy relationships.Biochim Biophys Acta. 2015 Oct;1850(10):2048-60. doi: 10.1016/j.bbagen.2015.01.013. Epub 2015 Jan 29. Biochim Biophys Acta. 2015. PMID: 25641576 Review.
-
KeePin' the p53 family in good shape.Cell Cycle. 2004 Jul;3(7):905-11. Epub 2004 Jul 2. Cell Cycle. 2004. PMID: 15254434 Review.
Cited by
-
miR-663 sustains NSCLC by inhibiting mitochondrial outer membrane permeabilization (MOMP) through PUMA/BBC3 and BTG2.Cell Death Dis. 2018 Jan 19;9(2):49. doi: 10.1038/s41419-017-0080-x. Cell Death Dis. 2018. PMID: 29352138 Free PMC article.
-
[Rita induce acute lymphoblostic leukemia cell apoptosis by activating P53 pathway].Zhonghua Xue Ye Xue Za Zhi. 2017 Feb 14;38(2):160-163. doi: 10.3760/cma.j.issn.0253-2727.2017.02.015. Zhonghua Xue Ye Xue Za Zhi. 2017. PMID: 28279043 Free PMC article. Chinese. No abstract available.
-
p53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner.Proc Natl Acad Sci U S A. 2015 Feb 10;112(6):1779-84. doi: 10.1073/pnas.1410723112. Epub 2015 Jan 26. Proc Natl Acad Sci U S A. 2015. PMID: 25624484 Free PMC article.
-
Decrease of mitochondrial p53 during late apoptosis is linked to its dephosphorylation on serine 20.Cancer Biol Ther. 2015;16(9):1296-307. doi: 10.1080/15384047.2015.1070978. Cancer Biol Ther. 2015. PMID: 26252178 Free PMC article.
-
Forecasting cell death dose-response from early signal transduction responses in vitro.Toxicol Sci. 2014 Aug 1;140(2):338-51. doi: 10.1093/toxsci/kfu089. Epub 2014 May 13. Toxicol Sci. 2014. PMID: 24824809 Free PMC article.
References
-
- Talos F, Petrenko O, Mena P, Moll UM. Mitochondrially targeted p53 has tumor suppressor activities in vivo. Cancer Res. 2005;65:9971–9981. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

