Glutamine depletion and glucose depletion trigger growth inhibition via distinctive gene expression reprogramming
- PMID: 22935705
- PMCID: PMC3478318
- DOI: 10.4161/cc.21944
Glutamine depletion and glucose depletion trigger growth inhibition via distinctive gene expression reprogramming
Abstract
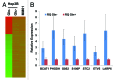
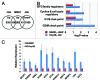
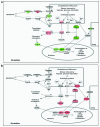
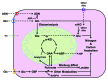
Glutamine (Gln) and glucose (Glc) represent two important nutrients for proliferating cells, consistent with the observations that oncogenic processes are associated with enhanced glycolysis and glutaminolysis. Gln depletion and Glc depletion have been shown to trigger growth arrest and eventually cell death. Solid tumors often outgrow the blood supply, resulting in ischemia, which is associated with hypoxia and nutrient insufficiency. Whereas oxygen-sensing and adaptive mechanisms to hypoxia have been well-studied, how cells directly sense and respond to Gln and Glc insufficiency remains unclear. Using mRNA profiling techniques, we compared the gene expression profiles of acute Gln-depleted cells, Glc-depleted cells and cells adapted to Gln depletion. Here we report the global changes of the gene expression in those cells cultured under the defined nutrient conditions. Analysis of mRNA profiling data revealed that Gln and Glc depletion triggered dramatic gene expression reprogramming. Either Gln or Glc deletion leads to changes of the expression of cell cycle genes, but these conditions have distinctive effects on transcription regulators and gene expression profiles. Moreover, Gln and Glc depletion triggered distinguishable ER-stress responses. The gene expression patterns support that Gln and Glc have distinctive metabolic roles in supporting cell survival and proliferation, and cells use different mechanisms to sense and respond to Gln and Glc insufficiency. Our mRNA profiling database provides a resource for further investigating the nutrient-sensing mechanisms and potential effects of Glc and Gln abundance on the biological behaviors of cells.
Figures

References
-
- Sang N, Fang J, Srinivas V, Leshchinsky I, Caro J. Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1 alpha is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol Cell Biol. 2002;22:2984–92. doi: 10.1128/MCB.22.9.2984-2992.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
