Kruppel-like factor 7 overexpression suppresses hematopoietic stem and progenitor cell function
- PMID: 22936656
- PMCID: PMC3471512
- DOI: 10.1182/blood-2012-02-409839
Kruppel-like factor 7 overexpression suppresses hematopoietic stem and progenitor cell function
Abstract
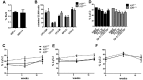
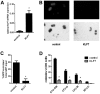
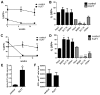
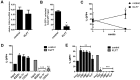
Increased expression of Kruppel-like factor 7 (KLF7) is an independent predictor of poor outcome in pediatric acute lymphoblastic leukemia. The contribution of KLF7 to hematopoiesis has not been previously described. Herein, we characterized the effect on murine hematopoiesis of the loss of KLF7 and enforced expression of KLF7. Long-term multilineage engraftment of Klf7(-/-) cells was comparable with control cells, and self-renewal, as assessed by serial transplantation, was not affected. Enforced expression of KLF7 results in a marked suppression of myeloid progenitor cell growth and a loss of short- and long-term repopulating activity. Interestingly, enforced expression of KLF7, although resulting in multilineage growth suppression that extended to hematopoietic stem cells and common lymphoid progenitors, spared T cells and enhanced the survival of early thymocytes. RNA expression profiling of KLF7-overexpressing hematopoietic progenitors identified several potential target genes mediating these effects. Notably, the known KLF7 target Cdkn1a (p21(Cip1/Waf1)) was not induced by KLF7, and loss of CDKN1A does not rescue the repopulating defect. These results suggest that KLF7 is not required for normal hematopoietic stem and progenitor function, but increased expression, as seen in a subset of lymphoid leukemia, inhibits myeloid cell proliferation and promotes early thymocyte survival.
Figures


References
-
- Coustan-Smith E, Sancho J, Behm FG, et al. Prognostic importance of measuring early clearance of leukemic cells by flow cytometry in childhood acute lymphoblastic leukemia. Blood. 2002;100(1):52–58. - PubMed
-
- Cammarata G, Augugliaro L, La Rosa M, et al. Gene expression profile of chronic myeloid leukemia innately resistant to Imatinib. Clin Leukemia. 2007;1(4):234–242.
-
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375(6529):316–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

