Chromatin and epigenetic regulation of pre-mRNA processing
- PMID: 22936691
- PMCID: PMC3459648
- DOI: 10.1093/hmg/dds353
Chromatin and epigenetic regulation of pre-mRNA processing
Abstract
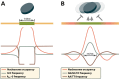
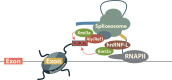
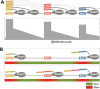
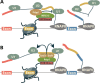
New data are revealing a complex landscape of gene regulation shaped by chromatin states that extend into the bodies of transcribed genes and associate with distinct RNA elements such as exons, introns and polyadenylation sites. Exons are characterized by increased levels of nucleosome positioning, DNA methylation and certain histone modifications. As pre-mRNA splicing occurs co-transcriptionally, changes in the transcription elongation rate or epigenetic marks can influence exon splicing. These new discoveries broaden our understanding of the epigenetic code and ascribe a novel role for chromatin in controlling pre-mRNA processing. In this review, we summarize the recently discovered interplay between the modulation of chromatin states and pre-mRNA processing with the particular focus on how these processes communicate with one another to control gene expression.
Figures
Similar articles
-
Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast.Genome Biol. 2011 Aug 22;12(8):R82. doi: 10.1186/gb-2011-12-8-r82. Genome Biol. 2011. PMID: 21859475 Free PMC article.
-
Chromatin's thread to alternative splicing regulation.Chromosoma. 2013 Dec;122(6):465-74. doi: 10.1007/s00412-013-0425-x. Epub 2013 Aug 3. Chromosoma. 2013. PMID: 23912688 Review.
-
Chromatin and splicing.Methods Mol Biol. 2014;1126:97-113. doi: 10.1007/978-1-62703-980-2_7. Methods Mol Biol. 2014. PMID: 24549658 Free PMC article.
-
Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms.Nucleic Acids Res. 2014 Jan;42(2):701-13. doi: 10.1093/nar/gkt875. Epub 2013 Sep 29. Nucleic Acids Res. 2014. PMID: 24081581 Free PMC article. Review.
-
A new link between transcriptional initiation and pre-mRNA splicing: The RNA binding histone variant H2A.B.PLoS Genet. 2017 Feb 24;13(2):e1006633. doi: 10.1371/journal.pgen.1006633. eCollection 2017 Feb. PLoS Genet. 2017. PMID: 28234895 Free PMC article.
Cited by
-
H3K4me3 demethylation by the histone demethylase KDM5C/JARID1C promotes DNA replication origin firing.Nucleic Acids Res. 2015 Mar 11;43(5):2560-74. doi: 10.1093/nar/gkv090. Epub 2015 Feb 23. Nucleic Acids Res. 2015. PMID: 25712104 Free PMC article.
-
TINTIN, at the interface of chromatin, transcription elongation, and mRNA processing.RNA Biol. 2015;12(5):486-9. doi: 10.1080/15476286.2015.1026032. RNA Biol. 2015. PMID: 25775193 Free PMC article.
-
The chromatin remodeler ZmCHB101 impacts alternative splicing contexts in response to osmotic stress.Plant Cell Rep. 2019 Feb;38(2):131-145. doi: 10.1007/s00299-018-2354-x. Epub 2018 Nov 15. Plant Cell Rep. 2019. PMID: 30443733
-
SRSF2-p95 hotspot mutation is highly associated with advanced forms of mastocytosis and mutations in epigenetic regulator genes.Haematologica. 2014 May;99(5):830-5. doi: 10.3324/haematol.2013.095133. Epub 2014 Jan 3. Haematologica. 2014. PMID: 24389310 Free PMC article.
-
A reassessment of DNA-immunoprecipitation-based genomic profiling.Nat Methods. 2018 Jul;15(7):499-504. doi: 10.1038/s41592-018-0038-7. Epub 2018 Jun 25. Nat Methods. 2018. PMID: 29941872 Free PMC article.
References
-
- Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi:10.1038/nature08909. - DOI - PMC - PubMed
-
- Di Giammartino D.C., Nishida K., Manley J.L. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi:10.1016/j.molcel.2011.08.017. - DOI - PMC - PubMed
-
- Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., BURGE C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi:10.1038/nature07509. - DOI - PMC - PubMed
-
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi:10.1038/ng.259. - DOI - PubMed
-
- Hertel K.J. Combinatorial control of exon recognition. J. Biol. Chem. 2008;283:1211–1215. doi:10.1074/jbc.R700035200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

