Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees
- PMID: 22936717
- PMCID: PMC3525142
- DOI: 10.1093/molbev/mss210
Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees
Abstract
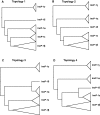
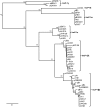
Plasmids of the incompatibility group IncP-1 can transfer and replicate in many genera of the Proteobacteria. They are composed of backbone genes that encode a variety of essential functions and accessory genes that have implications for human health and environmental remediation. Although it is well understood that the accessory genes are transferred horizontally between plasmids, recent studies have also provided examples of recombination in the backbone genes of IncP-1 plasmids. As a consequence, phylogeny estimation based on backbone genes is expected to produce conflicting gene tree topologies. The main goal of this study was therefore to infer the evolutionary history of IncP-1 plasmids in the presence of both vertical and horizontal gene transfer. This was achieved by quantifying the incongruence among gene trees and attributing it to known causes such as 1) phylogenetic uncertainty, 2) coalescent stochasticity, and 3) horizontal inheritance. Topologies of gene trees exhibited more incongruence than could be attributed to phylogenetic uncertainty alone. Species-tree estimation using a Bayesian framework that takes coalescent stochasticity into account was well supported, but it differed slightly from the maximum-likelihood tree estimated by concatenation of backbone genes. After removal of the gene that demonstrated a signal of intergroup recombination, the concatenated tree was congruent with the species-tree estimate, which itself was robust to inclusion/exclusion of the recombinant gene. Thus, in spite of horizontal gene exchange both within and among IncP-1 subgroups, the backbone genome of these IncP-1 plasmids retains a detectable vertical evolutionary history.
Figures



Similar articles
-
Diversification of broad host range plasmids correlates with the presence of antibiotic resistance genes.FEMS Microbiol Ecol. 2016 Jan;92(1):fiv151. doi: 10.1093/femsec/fiv151. Epub 2015 Dec 2. FEMS Microbiol Ecol. 2016. PMID: 26635412 Free PMC article.
-
Comparative genomics of pAKD4, the prototype IncP-1delta plasmid with a complete backbone.Plasmid. 2010 Mar;63(2):98-107. doi: 10.1016/j.plasmid.2009.11.005. Epub 2009 Dec 16. Plasmid. 2010. PMID: 20018208 Free PMC article.
-
Diversity of IncP-9 plasmids of Pseudomonas.Microbiology (Reading). 2008 Oct;154(Pt 10):2929-2941. doi: 10.1099/mic.0.2008/017939-0. Microbiology (Reading). 2008. PMID: 18832300 Free PMC article.
-
Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool.FEMS Microbiol Rev. 2007 Jul;31(4):449-77. doi: 10.1111/j.1574-6976.2007.00074.x. Epub 2007 Jun 6. FEMS Microbiol Rev. 2007. PMID: 17553065 Review.
-
The evolution of IncP catabolic plasmids.Curr Opin Biotechnol. 2005 Jun;16(3):291-8. doi: 10.1016/j.copbio.2005.04.002. Curr Opin Biotechnol. 2005. PMID: 15961030 Review.
Cited by
-
Novel Self-Transmissible and Broad-Host-Range Plasmids Exogenously Captured From Anaerobic Granules or Cow Manure.Front Microbiol. 2018 Nov 6;9:2602. doi: 10.3389/fmicb.2018.02602. eCollection 2018. Front Microbiol. 2018. PMID: 30459733 Free PMC article.
-
Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids.Nat Commun. 2020 Jul 17;11(1):3602. doi: 10.1038/s41467-020-17278-2. Nat Commun. 2020. PMID: 32681114 Free PMC article.
-
Differentially Marked IncP-1β R751 Plasmids for Cloning via Recombineering and Conjugation.Pol J Microbiol. 2019 Dec;68(4):559-563. doi: 10.33073/pjm-2019-052. Epub 2019 Dec 5. Pol J Microbiol. 2019. PMID: 31880899 Free PMC article.
-
IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage.Antimicrob Agents Chemother. 2017 Jan 24;61(2):e02229-16. doi: 10.1128/AAC.02229-16. Print 2017 Feb. Antimicrob Agents Chemother. 2017. PMID: 27895009 Free PMC article.
-
Plant Species-Dependent Increased Abundance and Diversity of IncP-1 Plasmids in the Rhizosphere: New Insights Into Their Role and Ecology.Front Microbiol. 2020 Nov 27;11:590776. doi: 10.3389/fmicb.2020.590776. eCollection 2020. Front Microbiol. 2020. PMID: 33329469 Free PMC article.
References
-
- Adamczyk M, Jagura-Burdzy G. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim Pol. 2003;50:425–453. - PubMed
-
- Akiyama T, Asfahl KL, Savin MC. Broad-host-range plasmids in treated wastewater effluent and receiving streams. J Environ Qual. 2010;39:2211–2215. - PubMed
-
- Bahl MI, Hansen LH, Goesmann A, Sørensen SJ. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established α, β and δ sub-groups. Plasmid. 2007;58:31–43. - PubMed
-
- Binh CT, Heuer H, Kaupenjohann M, Smalla K. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol Ecol. 2008;66:25–37. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

