Prediction of disease-related interactions between microRNAs and environmental factors based on a semi-supervised classifier
- PMID: 22937049
- PMCID: PMC3427386
- DOI: 10.1371/journal.pone.0043425
Prediction of disease-related interactions between microRNAs and environmental factors based on a semi-supervised classifier
Abstract
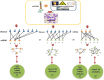
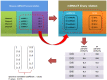
Accumulated evidence has shown that microRNAs (miRNAs) can functionally interact with a number of environmental factors (EFs) and their interactions critically affect phenotypes and diseases. Therefore, in-silico inference of disease-related miRNA-EF interactions is becoming crucial not only for the understanding of the mechanisms by which miRNAs and EFs contribute to disease, but also for disease diagnosis, treatment, and prognosis. In this paper, we analyzed the human miRNA-EF interaction data and revealed that miRNAs (EFs) with similar functions tend to interact with similar EFs (miRNAs) in the context of a given disease, which suggests a potential way to expand the current relation space of miRNAs, EFs, and diseases. Based on this observation, we further proposed a semi-supervised classifier based method (miREFScan) to predict novel disease-related interactions between miRNAs and EFs. As a result, the leave-one-out cross validation has shown that miREFScan obtained an AUC of 0.9564, indicating that miREFScan has a reliable performance. Moreover, we applied miREFScan to predict acute promyelocytic leukemia-related miRNA-EF interactions. The result shows that forty-nine of the top 1% predictions have been confirmed by experimental literature. In addition, using miREFScan we predicted and publicly released novel miRNA-EF interactions for 97 human diseases. Finally, we believe that miREFScan would be a useful bioinformatic resource for the research about the relationships among miRNAs, EFs, and human diseases.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

