Circular permutation prediction reveals a viable backbone disconnection for split proteins: an approach in identifying a new functional split intein
- PMID: 22937103
- PMCID: PMC3427171
- DOI: 10.1371/journal.pone.0043820
Circular permutation prediction reveals a viable backbone disconnection for split proteins: an approach in identifying a new functional split intein
Abstract
Split-protein systems have emerged as a powerful tool for detecting biomolecular interactions and reporting biological reactions. However, reliable methods for identifying viable split sites are still unavailable. In this study, we demonstrated the feasibility that valid circular permutation (CP) sites in proteins have the potential to act as split sites and that CP prediction can be used to search for internal permissive sites for creating new split proteins. Using a protein ligase, intein, as a model, CP predictor facilitated the creation of circular permutants in which backbone opening imposes the least detrimental effects on intein folding. We screened a series of predicted intein CPs and identified stable and native-fold CPs. When the valid CP sites were introduced as split sites, there was a reduction in folding enthalpy caused by the new backbone opening; however, the coincident loss in entropy was sufficient to be compensated, yielding a favorable free energy for self-association. Since split intein is exploited in protein semi-synthesis, we tested the related protein trans-splicing (PTS) activities of the corresponding split inteins. Notably, a novel functional split intein composed of the N-terminal 36 residues combined with the remaining C-terminal fragment was identified. Its PTS activity was shown to be better than current reported two-piece intein with a short N-terminal segment. Thus, the incorporation of in silico CP prediction facilitated the design of split intein as well as circular permutants.
Conflict of interest statement
Figures



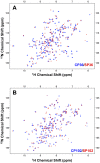
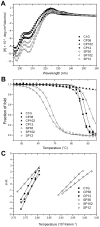


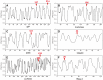
References
-
- Yu Y, Lutz S (2011) Circular permutation: a different way to engineer enzyme structure and function. Trends in biotechnology 29: 18–25. - PubMed
-
- Heinemann U, Hahn M (1995) Circular permutation of polypeptide chains: implications for protein folding and stability. Progress in biophysics and molecular biology 64: 121–143. - PubMed
-
- Lindqvist Y, Schneider G (1997) Circular permutations of natural protein sequences: structural evidence. Current opinion in structural biology 7: 422–427. - PubMed
-
- Whitehead TA, Bergeron LM, Clark DS (2009) Tying up the loose ends: circular permutation decreases the proteolytic susceptibility of recombinant proteins. Protein engineering, design & selection : PEDS 22: 607–613. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

