Crystal structures of archaemetzincin reveal a moldable substrate-binding site
- PMID: 22937112
- PMCID: PMC3427221
- DOI: 10.1371/journal.pone.0043863
Crystal structures of archaemetzincin reveal a moldable substrate-binding site
Abstract
Background: Archaemetzincins are metalloproteases occurring in archaea and some mammalia. They are distinct from all the other metzincins by their extended active site consensus sequence HEXXHXXGXXHCX(4)CXMX(17)CXXC featuring four conserved cysteine residues. Very little is known about their biological importance and structure-function relationships.
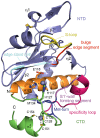
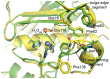
Principal findings: Here we present three crystal structures of the archaemetzincin AfAmzA (Uniprot O29917) from Archaeoglobus fulgidus, revealing a metzincin architecture featuring a zinc finger-like structural element involving the conserved cysteines of the consensus motif. The active sites in all three structures are occluded to different extents rendering the enzymes proteolytically inactive against a large variety of tested substrates. Owing to the different ligand binding there are significant differences in active site architecture, revealing a large flexibility of the loops covering the active site cleft.
Conclusions: The crystal structures of AfAmzA provide the structural basis for the lack of activity in standard proteolytic assays and imply a triggered activity onset upon opening of the active site cleft.
Conflict of interest statement
Figures
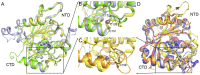


Similar articles
-
Architecture and function of metallopeptidase catalytic domains.Protein Sci. 2014 Feb;23(2):123-44. doi: 10.1002/pro.2400. Protein Sci. 2014. PMID: 24596965 Free PMC article. Review.
-
Identification and characterization of human archaemetzincin-1 and -2, two novel members of a family of metalloproteases widely distributed in Archaea.J Biol Chem. 2005 Aug 26;280(34):30367-75. doi: 10.1074/jbc.M504533200. Epub 2005 Jun 22. J Biol Chem. 2005. Retraction in: J Biol Chem. 2019 Jan 25;294(4):1434. doi: 10.1074/jbc.W118.007328. PMID: 15972818 Retracted.
-
Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus.J Mol Biol. 2007 Jun 29;370(1):128-41. doi: 10.1016/j.jmb.2007.04.050. Epub 2007 May 4. J Mol Biol. 2007. PMID: 17512006
-
Crystal structure of archaeal highly thermostable L-aspartate dehydrogenase/NAD/citrate ternary complex.FEBS J. 2007 Aug;274(16):4315-25. doi: 10.1111/j.1742-4658.2007.05961.x. Epub 2007 Jul 25. FEBS J. 2007. PMID: 17651440
-
Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains.Protein Sci. 2006 Aug;15(8):1815-28. doi: 10.1110/ps.052069306. Protein Sci. 2006. PMID: 16877706 Free PMC article. Review.
Cited by
-
A novel mechanism of latency in matrix metalloproteinases.J Biol Chem. 2015 Feb 20;290(8):4728-4740. doi: 10.1074/jbc.M114.605956. Epub 2015 Jan 2. J Biol Chem. 2015. PMID: 25555916 Free PMC article.
-
Architecture and function of metallopeptidase catalytic domains.Protein Sci. 2014 Feb;23(2):123-44. doi: 10.1002/pro.2400. Protein Sci. 2014. PMID: 24596965 Free PMC article. Review.
-
A Zinc-Dependent Protease AMZ-tk from a Thermophilic Archaeon is a New Member of the Archaemetzincin Protein Family.Front Microbiol. 2015 Dec 17;6:1380. doi: 10.3389/fmicb.2015.01380. eCollection 2015. Front Microbiol. 2015. PMID: 26733945 Free PMC article.
-
New mini- zincin structures provide a minimal scaffold for members of this metallopeptidase superfamily.BMC Bioinformatics. 2014 Jan 3;15:1. doi: 10.1186/1471-2105-15-1. BMC Bioinformatics. 2014. PMID: 24383880 Free PMC article.
References
-
- Jiang W, Bond JS (1992) Families of metalloendopeptidases and their relationships. FEBS Letters 312: 110–114. - PubMed
-
- Matthews BW (1988) Structural basis of the action of thermolysin and related zinc peptidases. Accounts of Chemical Research 21: 333–340.
-
- Gomis-Rüth FX (2003) Structural Aspects of the Metzincin Clan of Metalloendopeptidases. Molecular Biotechnology 24. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

