Sequencing and analysis of the gastrula transcriptome of the brittle star Ophiocoma wendtii
- PMID: 22938175
- PMCID: PMC3492025
- DOI: 10.1186/2041-9139-3-19
Sequencing and analysis of the gastrula transcriptome of the brittle star Ophiocoma wendtii
Abstract
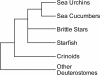
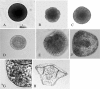
Background: The gastrula stage represents the point in development at which the three primary germ layers diverge. At this point the gene regulatory networks that specify the germ layers are established and the genes that define the differentiated states of the tissues have begun to be activated. These networks have been well-characterized in sea urchins, but not in other echinoderms. Embryos of the brittle star Ophiocoma wendtii share a number of developmental features with sea urchin embryos, including the ingression of mesenchyme cells that give rise to an embryonic skeleton. Notable differences are that no micromeres are formed during cleavage divisions and no pigment cells are formed during development to the pluteus larval stage. More subtle changes in timing of developmental events also occur. To explore the molecular basis for the similarities and differences between these two echinoderms, we have sequenced and characterized the gastrula transcriptome of O. wendtii.
Methods: Development of Ophiocoma wendtii embryos was characterized and RNA was isolated from the gastrula stage. A transcriptome data base was generated from this RNA and was analyzed using a variety of methods to identify transcripts expressed and to compare those transcripts to those expressed at the gastrula stage in other organisms.
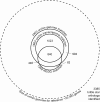
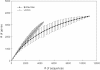
Results: Using existing databases, we identified brittle star transcripts that correspond to 3,385 genes, including 1,863 genes shared with the sea urchin Strongylocentrotus purpuratus gastrula transcriptome. We characterized the functional classes of genes present in the transcriptome and compared them to those found in this sea urchin. We then examined those members of the germ-layer specific gene regulatory networks (GRNs) of S. purpuratus that are expressed in the O. wendtii gastrula. Our results indicate that there is a shared 'genetic toolkit' central to the echinoderm gastrula, a key stage in embryonic development, though there are also differences that reflect changes in developmental processes.
Conclusions: The brittle star expresses genes representing all functional classes at the gastrula stage. Brittle stars and sea urchins have comparable numbers of each class of genes and share many of the genes expressed at gastrulation. Examination of the brittle star genes in which sea urchin orthologs are utilized in germ layer specification reveals a relatively higher level of conservation of key regulatory components compared to the overall transcriptome. We also identify genes that were either lost or whose temporal expression has diverged from that of sea urchins.
Figures


Similar articles
-
Examination of the skeletal proteome of the brittle star Ophiocoma wendtii reveals overall conservation of proteins but variation in spicule matrix proteins.Proteome Sci. 2015 Feb 7;13:7. doi: 10.1186/s12953-015-0064-7. eCollection 2015. Proteome Sci. 2015. PMID: 25705131 Free PMC article.
-
Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution.Genome Biol. 2018 Feb 28;19(1):26. doi: 10.1186/s13059-018-1402-8. Genome Biol. 2018. PMID: 29490679 Free PMC article.
-
The skeletal proteome of the sea star Patiria miniata and evolution of biomineralization in echinoderms.BMC Evol Biol. 2017 Jun 5;17(1):125. doi: 10.1186/s12862-017-0978-z. BMC Evol Biol. 2017. PMID: 28583083 Free PMC article.
-
Developmental gene regulatory network evolution: insights from comparative studies in echinoderms.Genesis. 2014 Mar;52(3):193-207. doi: 10.1002/dvg.22757. Epub 2014 Mar 6. Genesis. 2014. PMID: 24549884 Review.
-
Gastrulation in the sea urchin.Curr Top Dev Biol. 2020;136:195-218. doi: 10.1016/bs.ctdb.2019.08.004. Epub 2019 Oct 22. Curr Top Dev Biol. 2020. PMID: 31959288 Free PMC article. Review.
Cited by
-
Cell type phylogenetics informs the evolutionary origin of echinoderm larval skeletogenic cell identity.Commun Biol. 2019 May 3;2:160. doi: 10.1038/s42003-019-0417-3. eCollection 2019. Commun Biol. 2019. PMID: 31069269 Free PMC article.
-
Phylogenomic analyses of Echinodermata support the sister groups of Asterozoa and Echinozoa.PLoS One. 2015 Mar 20;10(3):e0119627. doi: 10.1371/journal.pone.0119627. eCollection 2015. PLoS One. 2015. PMID: 25794146 Free PMC article.
-
Examination of the skeletal proteome of the brittle star Ophiocoma wendtii reveals overall conservation of proteins but variation in spicule matrix proteins.Proteome Sci. 2015 Feb 7;13:7. doi: 10.1186/s12953-015-0064-7. eCollection 2015. Proteome Sci. 2015. PMID: 25705131 Free PMC article.
-
Gene expression profiling during the embryo-to-larva transition in the giant red sea urchin Mesocentrotus franciscanus.Ecol Evol. 2017 Mar 21;7(8):2798-2811. doi: 10.1002/ece3.2850. eCollection 2017 Apr. Ecol Evol. 2017. PMID: 28428870 Free PMC article.
-
Branching out: origins of the sea urchin larval skeleton in development and evolution.Genesis. 2014 Mar;52(3):173-85. doi: 10.1002/dvg.22756. Epub 2014 Mar 5. Genesis. 2014. PMID: 24549853 Free PMC article. Review.
References
-
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. - DOI - PubMed
-
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Schilstra MJ, Clarke PJ, Rust AG, Pan Z, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. - DOI - PubMed
-
- Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev. 2004;204:351–380. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

