Appraising the performance of genotyping tools in the prediction of coreceptor tropism in HIV-1 subtype C viruses
- PMID: 22938574
- PMCID: PMC3482586
- DOI: 10.1186/1471-2334-12-203
Appraising the performance of genotyping tools in the prediction of coreceptor tropism in HIV-1 subtype C viruses
Abstract
Background: In human immunodeficiency virus type 1 (HIV-1) infection, transmitted viruses generally use the CCR5 chemokine receptor as a coreceptor for host cell entry. In more than 50% of subtype B infections, a switch in coreceptor tropism from CCR5- to CXCR4-use occurs during disease progression. Phenotypic or genotypic approaches can be used to test for the presence of CXCR4-using viral variants in an individual's viral population that would result in resistance to treatment with CCR5-antagonists. While genotyping approaches for coreceptor-tropism prediction in subtype B are well established and verified, they are less so for subtype C.
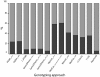
Methods: Here, using a dataset comprising V3 loop sequences from 349 CCR5-using and 56 CXCR4-using HIV-1 subtype C viruses we perform a comparative analysis of the predictive ability of 11 genotypic algorithms in their prediction of coreceptor tropism in subtype C. We calculate the sensitivity and specificity of each of the approaches as well as determining their overall accuracy. By separating the CXCR4-using viruses into CXCR4-exclusive (25 sequences) and dual-tropic (31 sequences) we evaluate the effect of the possible conflicting signal from dual-tropic viruses on the ability of a of the approaches to correctly predict coreceptor phenotype.
Results: We determined that geno2pheno with a false positive rate of 5% is the best approach for predicting CXCR4-usage in subtype C sequences with an accuracy of 94% (89% sensitivity and 99% specificity). Contrary to what has been reported for subtype B, the optimal approaches for prediction of CXCR4-usage in sequence from viruses that use CXCR4 exclusively, also perform best at predicting CXCR4-use in dual-tropic viral variants.
Conclusions: The accuracy of genotyping approaches at correctly predicting the coreceptor usage of V3 sequences from subtype C viruses is very high. We suggest that genotyping approaches can be used to test for coreceptor tropism in HIV-1 group M subtype C with a high degree of confidence that they will identify CXCR4-usage in both CXCR4-exclusive and dual tropic variants.
Figures
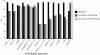
Similar articles
-
HIV-1 tropism determination using a phenotypic Env recombinant viral assay highlights overestimation of CXCR4-usage by genotypic prediction algorithms for CRF01_AE and CRF02_AG [corrected].PLoS One. 2013 May 8;8(5):e60566. doi: 10.1371/journal.pone.0060566. Print 2013. PLoS One. 2013. PMID: 23667426 Free PMC article.
-
Genotypic prediction of HIV-1 subtype D tropism.Retrovirology. 2011 Jul 13;8:56. doi: 10.1186/1742-4690-8-56. Retrovirology. 2011. PMID: 21752271 Free PMC article.
-
Frequent CXCR4 tropism of HIV-1 subtype A and CRF02_AG during late-stage disease--indication of an evolving epidemic in West Africa.Retrovirology. 2010 Mar 22;7:23. doi: 10.1186/1742-4690-7-23. Retrovirology. 2010. PMID: 20307309 Free PMC article.
-
HIV-1 subtype C predicted co-receptor tropism in Africa: an individual sequence level meta-analysis.AIDS Res Ther. 2020 Feb 7;17(1):5. doi: 10.1186/s12981-020-0263-x. AIDS Res Ther. 2020. PMID: 32033571 Free PMC article. Review.
-
Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
Cited by
-
Features of Recently Transmitted HIV-1 Clade C Viruses that Impact Antibody Recognition: Implications for Active and Passive Immunization.PLoS Pathog. 2016 Jul 19;12(7):e1005742. doi: 10.1371/journal.ppat.1005742. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27434311 Free PMC article.
-
Risk of breast cancer with CXCR4-using HIV defined by V3 loop sequencing.J Acquir Immune Defic Syndr. 2015 Jan 1;68(1):30-5. doi: 10.1097/QAI.0000000000000400. J Acquir Immune Defic Syndr. 2015. PMID: 25321183 Free PMC article.
-
Monophylogenetic HIV-1C epidemic in Ethiopia is dominated by CCR5-tropic viruses-an analysis of a prospective country-wide cohort.BMC Infect Dis. 2017 Jan 6;17(1):37. doi: 10.1186/s12879-016-2163-1. BMC Infect Dis. 2017. PMID: 28061826 Free PMC article.
-
Partial HIV C2V3 envelope sequence analysis reveals association of coreceptor tropism, envelope glycosylation and viral genotypic variability among Kenyan patients on HAART.Virol J. 2017 Feb 14;14(1):29. doi: 10.1186/s12985-017-0703-y. Virol J. 2017. PMID: 28196510 Free PMC article.
-
Prediction of coreceptor usage by five bioinformatics tools in a large Ethiopian HIV-1 subtype C cohort.PLoS One. 2017 Aug 25;12(8):e0182384. doi: 10.1371/journal.pone.0182384. eCollection 2017. PLoS One. 2017. PMID: 28841646 Free PMC article.
References
-
- McDougal JS, Maddon PJ, Dalgleish AG, Clapham PR, Littman DR, Godfrey M, Maddon DE, Chess L, Weiss RA, Axel R. The T4 glycoprotein is a cell-surface receptor for the AIDS virus. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 2):703–711. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

