Role of aldo-keto reductases and other doxorubicin pharmacokinetic genes in doxorubicin resistance, DNA binding, and subcellular localization
- PMID: 22938713
- PMCID: PMC3495881
- DOI: 10.1186/1471-2407-12-381
Role of aldo-keto reductases and other doxorubicin pharmacokinetic genes in doxorubicin resistance, DNA binding, and subcellular localization
Abstract
Background: Since proteins involved in chemotherapy drug pharmacokinetics and pharmacodynamics have a strong impact on the uptake, metabolism, and efflux of such drugs, they likely play critical roles in resistance to chemotherapy drugs in cancer patients.
Methods: To investigate this hypothesis, we conducted a whole genome microarray study to identify difference in the expression of genes between isogenic doxorubicin-sensitive and doxorubicin-resistant MCF-7 breast tumour cells. We then assessed the degree of over-representation of doxorubicin pharmacokinetic and pharmacodynamic genes in the dataset of doxorubicin resistance genes.
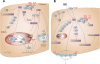
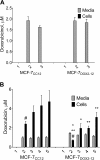
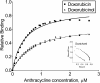
Results: Of 27,958 Entrez genes on the array, 7.4 per cent or 2,063 genes were differentially expressed by ≥ 2-fold between wildtype and doxorubicin-resistant cells. The false discovery rate was set at 0.01 and the minimum p value for significance for any gene within the "hit list" was 0.01. Seventeen and 43 per cent of doxorubicin pharmacokinetic genes were over-represented in the hit list, depending upon whether the gene name was identical or within the same gene family, respectively. The most over-represented genes were within the 1C and 1B families of aldo-keto reductases (AKRs), which convert doxorubicin to doxorubicinol. Other genes convert doxorubicin to other metabolites or affect the influx, efflux, or cytotoxicity of the drug. In further support of the role of AKRs in doxorubicin resistance, we observed that, in comparison to doxorubicin, doxorubincol exhibited dramatically reduced cytotoxicity, reduced DNA-binding activity, and strong localization to extra nuclear lysosomes. Pharmacologic inhibition of the above AKRs in doxorubicin-resistant cells increased cellular doxorubicin levels, restored doxorubicin cytotoxicity and re-established doxorubicin localization to the nucleus. The properties of doxorubicinol were unaffected.
Conclusions: These findings demonstrate the utility of using curated pharmacokinetic and pharmacodynamic knowledge bases to identify highly relevant genes associated with doxorubicin resistance. The induction of one or more of these genes was found to be correlated with changes in the drug's properties, while inhibiting one specific class of these genes (the AKRs) increased cellular doxorubicin content and restored drug DNA binding, cytotoxicity, and subcellular localization.
Figures

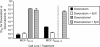

Similar articles
-
Proteasome inhibitors MG-132 and bortezomib induce AKR1C1, AKR1C3, AKR1B1, and AKR1B10 in human colon cancer cell lines SW-480 and HT-29.Chem Biol Interact. 2011 May 30;191(1-3):239-49. doi: 10.1016/j.cbi.2010.12.026. Epub 2011 Jan 6. Chem Biol Interact. 2011. PMID: 21215737
-
Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin.J Pharmacol Exp Ther. 2010 Dec;335(3):533-45. doi: 10.1124/jpet.110.173179. Epub 2010 Sep 13. J Pharmacol Exp Ther. 2010. PMID: 20837989
-
Alterations in estrogen signalling pathways upon acquisition of anthracycline resistance in breast tumor cells.PLoS One. 2017 Feb 14;12(2):e0172244. doi: 10.1371/journal.pone.0172244. eCollection 2017. PLoS One. 2017. PMID: 28196134 Free PMC article.
-
Aldo-keto reductases: Role in cancer development and theranostics.Oncol Res. 2024 Jul 17;32(8):1287-1308. doi: 10.32604/or.2024.049918. eCollection 2024. Oncol Res. 2024. PMID: 39055885 Free PMC article. Review.
-
Molecular determinants of steroid recognition and catalysis in aldo-keto reductases. Lessons from 3alpha-hydroxysteroid dehydrogenase.J Steroid Biochem Mol Biol. 1999 Apr-Jun;69(1-6):211-25. doi: 10.1016/s0960-0760(99)00038-2. J Steroid Biochem Mol Biol. 1999. PMID: 10418995 Review.
Cited by
-
A gravity-driven tissue chip to study the efficacy and toxicity of cancer therapeutics.Lab Chip. 2024 Nov 19;24(23):5251-5263. doi: 10.1039/d4lc00404c. Lab Chip. 2024. PMID: 39485368 Free PMC article.
-
PredictiveNetwork: predictive gene network estimation with application to gastric cancer drug response-predictive network analysis.BMC Bioinformatics. 2022 Aug 16;23(1):342. doi: 10.1186/s12859-022-04871-z. BMC Bioinformatics. 2022. PMID: 35974335 Free PMC article.
-
A novel selective AKR1C3-activated prodrug AST-3424/OBI-3424 exhibits broad anti-tumor activity.Am J Cancer Res. 2021 Jul 15;11(7):3645-3659. eCollection 2021. Am J Cancer Res. 2021. PMID: 34354865 Free PMC article.
-
Selective AKR1C3 Inhibitors Potentiate Chemotherapeutic Activity in Multiple Acute Myeloid Leukemia (AML) Cell Lines.ACS Med Chem Lett. 2016 Jun 22;7(8):774-9. doi: 10.1021/acsmedchemlett.6b00163. eCollection 2016 Aug 11. ACS Med Chem Lett. 2016. PMID: 27563402 Free PMC article.
-
Potent and Highly Selective Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors Act as Chemotherapeutic Potentiators in Acute Myeloid Leukemia and T-Cell Acute Lymphoblastic Leukemia.J Med Chem. 2019 Apr 11;62(7):3590-3616. doi: 10.1021/acs.jmedchem.9b00090. Epub 2019 Mar 25. J Med Chem. 2019. PMID: 30836001 Free PMC article.
References
-
- Schneider YJ, Baurain R, Zenebergh A, Trouet A. DNA-binding parameters of daunorubicin and doxorubicin in the conditions used for studying the interaction of anthracycline-DNA complexes with cells in vitro. Cancer Chemother Pharmacol. 1979;2:7–10. - PubMed
-
- Lopez M. Anthracyclines in the adjuvant treatment of breast carcinoma: thirty years later. Clin Ter. 2006;157:165–177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

