Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania
- PMID: 22939620
- PMCID: PMC3684241
- DOI: 10.1016/j.cell.2012.07.030
Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania
Abstract
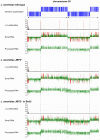
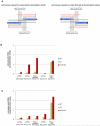
Some Ts in nuclear DNA of trypanosomes and Leishmania are hydroxylated and glucosylated to yield base J (β-D-glucosyl-hydroxymethyluracil). In Leishmania, about 99% of J is located in telomeric repeats. We show here that most of the remaining J is located at chromosome-internal RNA polymerase II termination sites. This internal J and telomeric J can be reduced by a knockout of J-binding protein 2 (JBP2), an enzyme involved in the first step of J biosynthesis. J levels are further reduced by growing Leishmania JBP2 knockout cells in BrdU-containing medium, resulting in cell death. The loss of internal J in JBP2 knockout cells is accompanied by massive readthrough at RNA polymerase II termination sites. The readthrough varies between transcription units but may extend over 100 kb. We conclude that J is required for proper transcription termination and infer that the absence of internal J kills Leishmania by massive readthrough of transcriptional stops.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

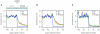


References
-
- Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. - PubMed
-
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, Raymond CK. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat. Methods. 2009;6:647–649. - PubMed
-
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 2011;12:123–135. - PubMed
-
- Belli SI, Monnerat S, Schaff C, Masina S, Noll T, Myler PJ, Stuart K, Fasel N. Sense and antisense transcripts in the histone H1 (HIS-1) locus of Leishmania major. Int. J. Parasitol. 2003;33:965–975. - PubMed
-
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

